Probe CUST_28162_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28162_PI426222305 | JHI_St_60k_v1 | DMT400004533 | CAATTAGATATGATTGATGGATGAAAAGATCCATAGAAAAGAAGCTCACGGTCATGATTC |
All Microarray Probes Designed to Gene DMG400001803
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28162_PI426222305 | JHI_St_60k_v1 | DMT400004533 | CAATTAGATATGATTGATGGATGAAAAGATCCATAGAAAAGAAGCTCACGGTCATGATTC |
| CUST_28145_PI426222305 | JHI_St_60k_v1 | DMT400004535 | CATTCGTCGCCCTGGGTAAAGCATCAAATTATTTCTGCAATTCAAACCTATAATTGATAA |
| CUST_28065_PI426222305 | JHI_St_60k_v1 | DMT400004534 | CAATTAGATATGATTGATGGATGAAAAGATCCATAGAAAAGAAGCTCACGGTCATGATTC |
Microarray Signals from CUST_28162_PI426222305
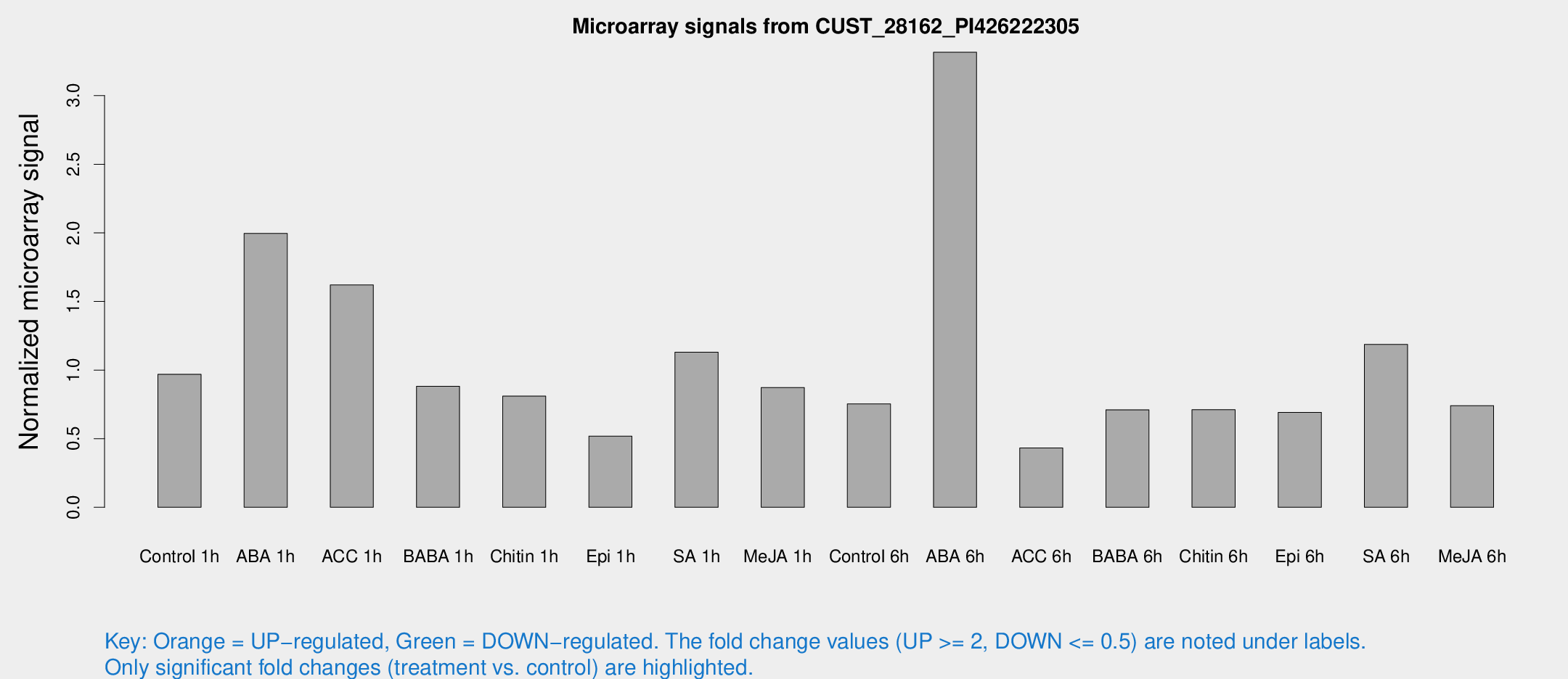
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14.7358 | 2.95773 | 0.969428 | 0.214209 |
| ABA 1h | 26.5026 | 4.32282 | 1.99545 | 0.41299 |
| ACC 1h | 24.5262 | 3.43987 | 1.62038 | 0.234963 |
| BABA 1h | 13.2624 | 3.49209 | 0.881571 | 0.24241 |
| Chitin 1h | 11.8623 | 3.29932 | 0.810299 | 0.326119 |
| Epi 1h | 7.02971 | 2.84837 | 0.518777 | 0.246366 |
| SA 1h | 18.418 | 5.08529 | 1.12967 | 0.321628 |
| Me-JA 1h | 11.3342 | 2.99983 | 0.872852 | 0.379995 |
| Control 6h | 12.6914 | 4.51771 | 0.753554 | 0.32217 |
| ABA 6h | 54.2633 | 11.0531 | 3.3161 | 0.834392 |
| ACC 6h | 8.18605 | 3.56855 | 0.432418 | 0.211446 |
| BABA 6h | 12.1751 | 3.35825 | 0.710727 | 0.212727 |
| Chitin 6h | 12.5624 | 4.21066 | 0.71156 | 0.30666 |
| Epi 6h | 13.2761 | 4.41849 | 0.692175 | 0.274535 |
| SA 6h | 17.5375 | 3.26429 | 1.18666 | 0.233772 |
| Me-JA 6h | 12.3418 | 4.08939 | 0.741017 | 0.309704 |
Source Transcript PGSC0003DMT400004533 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37240.1 | +2 | 3e-06 | 47 | 39/150 (26%) | Thioredoxin superfamily protein | chr2:15640235-15641720 REVERSE LENGTH=248 |
