Probe CUST_26406_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26406_PI426222305 | JHI_St_60k_v1 | DMT400037040 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
All Microarray Probes Designed to Gene DMG400014275
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26358_PI426222305 | JHI_St_60k_v1 | DMT400037041 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
| CUST_26406_PI426222305 | JHI_St_60k_v1 | DMT400037040 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
| CUST_26460_PI426222305 | JHI_St_60k_v1 | DMT400037042 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
Microarray Signals from CUST_26406_PI426222305
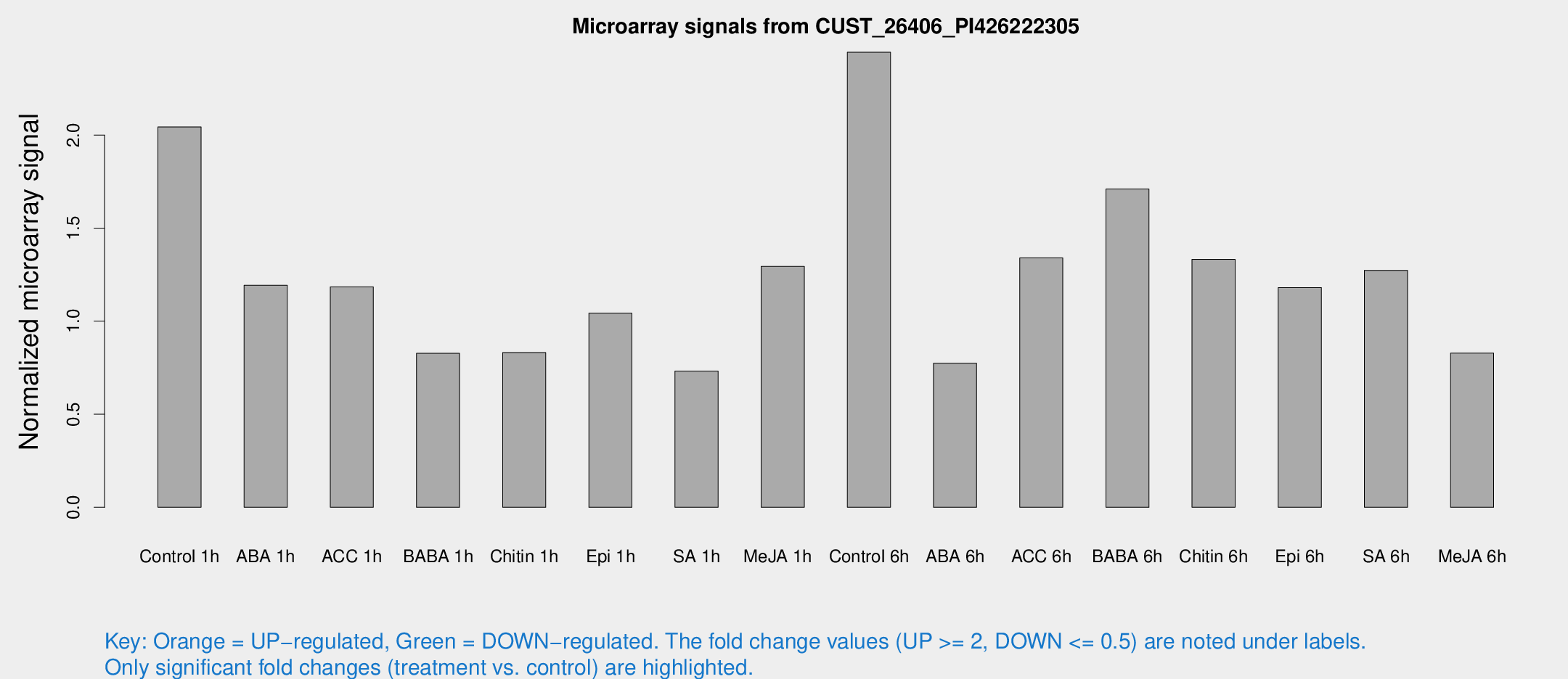
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19.5663 | 7.30809 | 2.04381 | 0.761236 |
| ABA 1h | 9.05584 | 3.53194 | 1.19361 | 0.503394 |
| ACC 1h | 10.9781 | 4.39507 | 1.1849 | 0.559258 |
| BABA 1h | 6.63588 | 3.84984 | 0.827293 | 0.479416 |
| Chitin 1h | 6.23894 | 3.6263 | 0.831616 | 0.481586 |
| Epi 1h | 8.06185 | 3.60192 | 1.04337 | 0.51807 |
| SA 1h | 6.25708 | 3.62944 | 0.732441 | 0.424614 |
| Me-JA 1h | 9.54509 | 3.81058 | 1.29467 | 0.615866 |
| Control 6h | 21.1737 | 4.34496 | 2.44557 | 0.515526 |
| ABA 6h | 6.84879 | 3.97591 | 0.774407 | 0.449191 |
| ACC 6h | 12.9964 | 4.64406 | 1.34035 | 0.470388 |
| BABA 6h | 29.7723 | 22.383 | 1.71062 | 2.15378 |
| Chitin 6h | 12.1345 | 4.30476 | 1.33291 | 0.502783 |
| Epi 6h | 11.8876 | 4.80281 | 1.18097 | 0.55643 |
| SA 6h | 10.9782 | 4.16871 | 1.27362 | 0.535065 |
| Me-JA 6h | 6.86419 | 3.82462 | 0.828723 | 0.462614 |
Source Transcript PGSC0003DMT400037040 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G45750.1 | +3 | 0.0 | 533 | 255/398 (64%) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | chr2:18842655-18845343 FORWARD LENGTH=631 |
