Probe CUST_26460_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26460_PI426222305 | JHI_St_60k_v1 | DMT400037042 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
All Microarray Probes Designed to Gene DMG400014275
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26358_PI426222305 | JHI_St_60k_v1 | DMT400037041 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
| CUST_26406_PI426222305 | JHI_St_60k_v1 | DMT400037040 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
| CUST_26460_PI426222305 | JHI_St_60k_v1 | DMT400037042 | GGAAAATTGATTAATCTTAAAGATGGTTCTATTAGAACCGCCATTGATACAGGATGTGGG |
Microarray Signals from CUST_26460_PI426222305
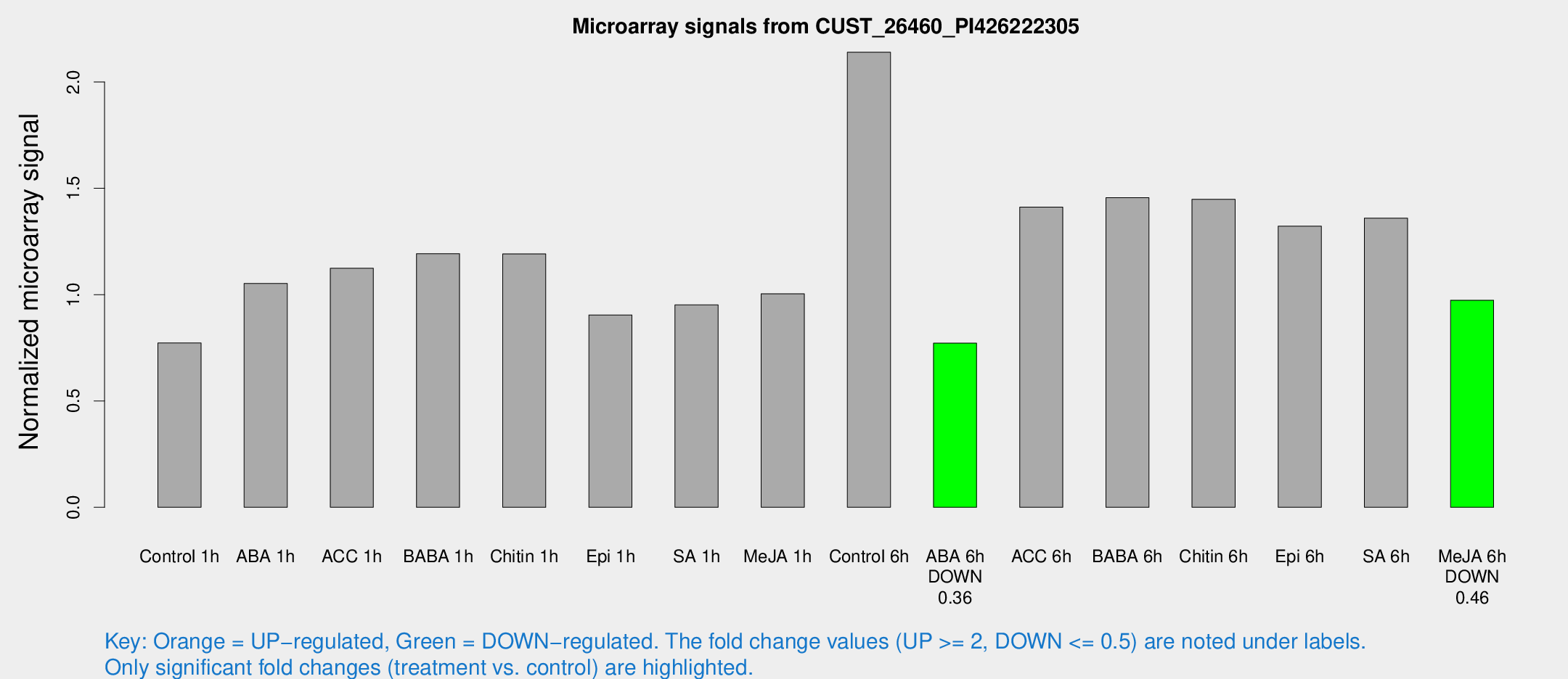
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4.94783 | 2.8734 | 0.772769 | 0.440826 |
| ABA 1h | 6.23663 | 2.77614 | 1.05249 | 0.507067 |
| ACC 1h | 8.43326 | 3.16326 | 1.12422 | 0.528936 |
| BABA 1h | 8.79008 | 3.69445 | 1.19265 | 0.557673 |
| Chitin 1h | 7.5121 | 2.91955 | 1.19162 | 0.539122 |
| Epi 1h | 4.96373 | 2.88187 | 0.904613 | 0.509814 |
| SA 1h | 6.77124 | 2.91007 | 0.951922 | 0.463105 |
| Me-JA 1h | 5.22978 | 2.94375 | 1.00365 | 0.550556 |
| Control 6h | 14.3614 | 3.08721 | 2.1399 | 0.498212 |
| ABA 6h | 5.34541 | 3.09902 | 0.77193 | 0.446389 |
| ACC 6h | 10.9736 | 3.66774 | 1.41163 | 0.535779 |
| BABA 6h | 11.3455 | 3.41466 | 1.45586 | 0.508477 |
| Chitin 6h | 10.2456 | 3.34367 | 1.44842 | 0.493341 |
| Epi 6h | 10.0452 | 3.47463 | 1.32197 | 0.514597 |
| SA 6h | 9.16256 | 3.18989 | 1.35996 | 0.534298 |
| Me-JA 6h | 6.36944 | 2.98761 | 0.973662 | 0.464856 |
Source Transcript PGSC0003DMT400037042 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G45750.1 | +3 | 0.0 | 870 | 415/627 (66%) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | chr2:18842655-18845343 FORWARD LENGTH=631 |
