Probe CUST_2542_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2542_PI426222305 | JHI_St_60k_v1 | DMT400072557 | GGCTTTCATGAATTTTAGAGGACTGTTTACTTTTTCTTGGAGATGCAATATGAAGGACAA |
All Microarray Probes Designed to Gene DMG400028231
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2184_PI426222305 | JHI_St_60k_v1 | DMT400072556 | CATCAGTAACTGATTCCGAGGCATCGCATGATGGTTGGAAAATGAAGGACAATGGTAACA |
| CUST_2542_PI426222305 | JHI_St_60k_v1 | DMT400072557 | GGCTTTCATGAATTTTAGAGGACTGTTTACTTTTTCTTGGAGATGCAATATGAAGGACAA |
Microarray Signals from CUST_2542_PI426222305
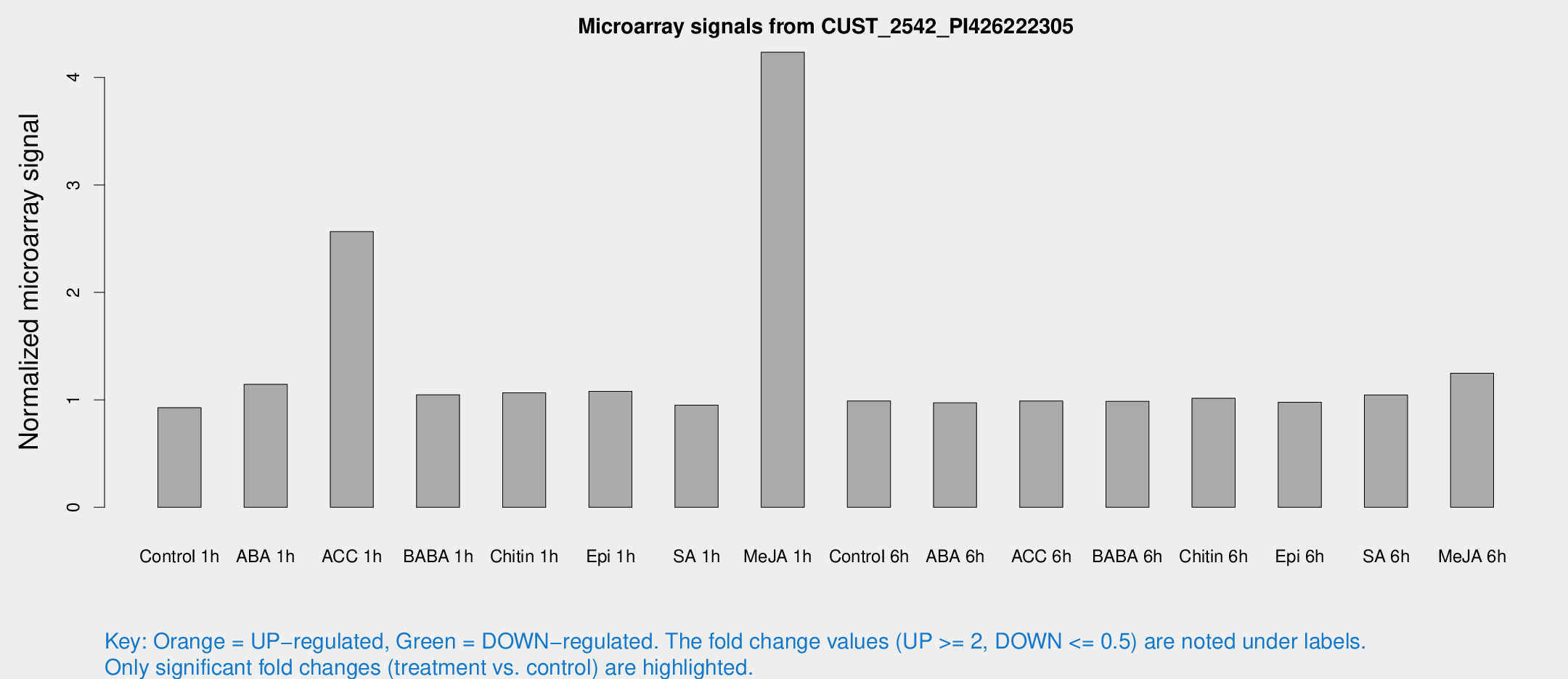
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.24016 | 3.04258 | 0.926597 | 0.533935 |
| ABA 1h | 5.81564 | 2.98592 | 1.14389 | 0.600832 |
| ACC 1h | 21.1647 | 11.8828 | 2.56575 | 2.52393 |
| BABA 1h | 5.7498 | 3.24285 | 1.04726 | 0.589711 |
| Chitin 1h | 5.35463 | 3.12083 | 1.06454 | 0.603664 |
| Epi 1h | 5.24043 | 3.04374 | 1.07955 | 0.61573 |
| SA 1h | 5.55919 | 3.07066 | 0.949813 | 0.526072 |
| Me-JA 1h | 19.7998 | 3.37618 | 4.23445 | 0.722925 |
| Control 6h | 5.6435 | 3.2702 | 0.989466 | 0.573034 |
| ABA 6h | 5.88774 | 3.35907 | 0.972147 | 0.554929 |
| ACC 6h | 6.5918 | 3.90818 | 0.989373 | 0.574018 |
| BABA 6h | 6.29375 | 3.65585 | 0.985614 | 0.57075 |
| Chitin 6h | 6.1439 | 3.56171 | 1.01444 | 0.587897 |
| Epi 6h | 6.33304 | 3.70621 | 0.978029 | 0.566591 |
| SA 6h | 5.8838 | 3.4081 | 1.04559 | 0.605482 |
| Me-JA 6h | 7.55997 | 3.29757 | 1.24761 | 0.612637 |
Source Transcript PGSC0003DMT400072557 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G06440.4 | +1 | 3e-116 | 374 | 207/439 (47%) | FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: mitochondrion; EXPRESSED IN: 22 plant structures; EXPRESSED DURING: 12 growth stages; BEST Arabidopsis thaliana protein match is: Polyketide cyclase/dehydrase and lipid transport superfamily protein (TAIR:AT3G11720.3). | chr5:1964641-1966807 REVERSE LENGTH=486 |
