Probe CUST_25375_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25375_PI426222305 | JHI_St_60k_v1 | DMT400034695 | AGACAGCGTGTCTAACGGCCTTTTGTTATAATCATTATAGTTCTTTCTTTGACTCTTCTT |
All Microarray Probes Designed to Gene DMG400013332
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25432_PI426222305 | JHI_St_60k_v1 | DMT400034698 | AAAGAAATGGCCACTCTATCTTAGCACAAAAAATACCATCCTTAAGAAATACGACGGAAG |
| CUST_25460_PI426222305 | JHI_St_60k_v1 | DMT400034694 | TGAAAGCGCTGAGGCTACTTTGAAATGACCCGTGTCATTTGGAAATTAATTAAGGATAAG |
| CUST_25291_PI426222305 | JHI_St_60k_v1 | DMT400034697 | TTTCACTAAAAATGGCTTTTCAAAAGATCAAAGTGGCTAACCCCATAGTTGAAATGGACG |
| CUST_25361_PI426222305 | JHI_St_60k_v1 | DMT400034696 | TAGCACTTATTATCCACGGATCCAAGATTTGGATCGCTTGGATTGATGACATCGATCCTG |
| CUST_25375_PI426222305 | JHI_St_60k_v1 | DMT400034695 | AGACAGCGTGTCTAACGGCCTTTTGTTATAATCATTATAGTTCTTTCTTTGACTCTTCTT |
Microarray Signals from CUST_25375_PI426222305
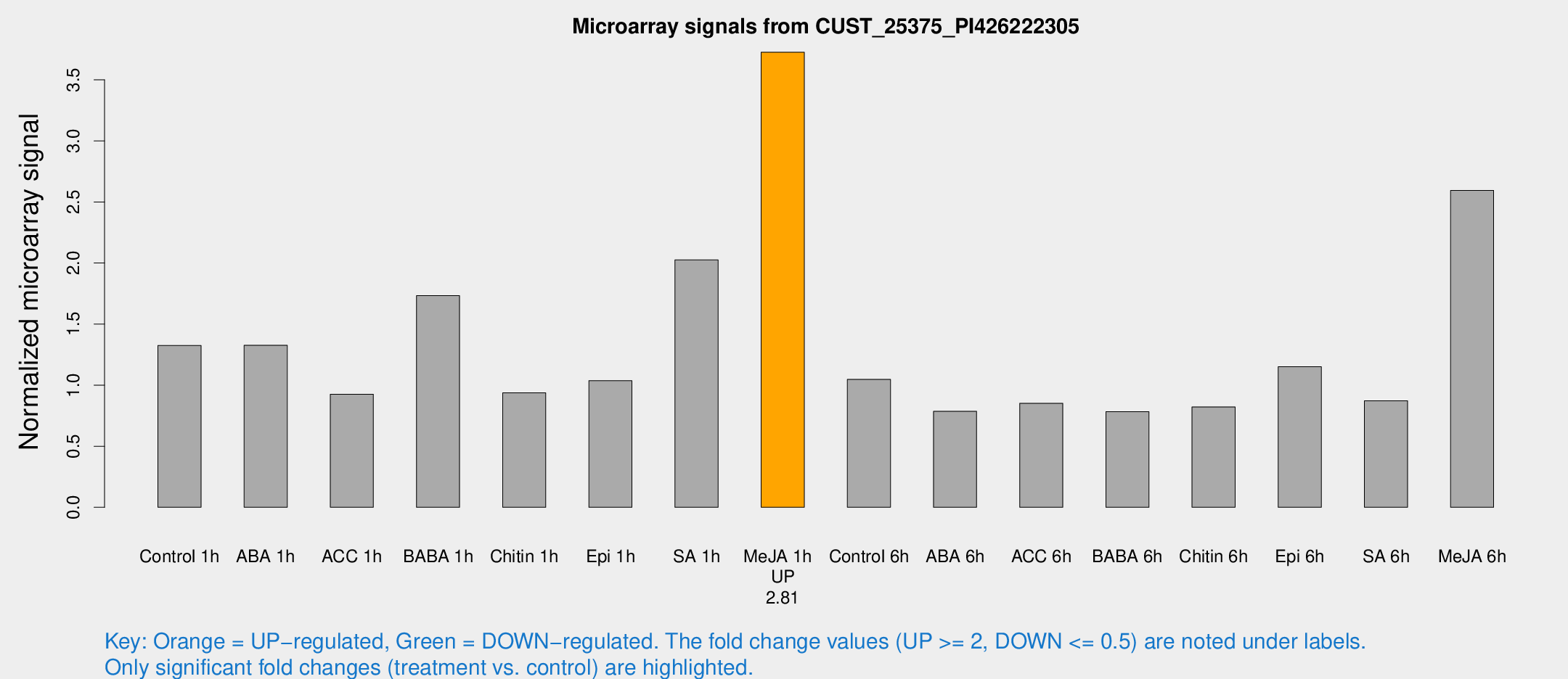
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.1736 | 3.13314 | 1.32534 | 0.3859 |
| ABA 1h | 10.092 | 3.07541 | 1.32667 | 0.447369 |
| ACC 1h | 8.57561 | 3.3983 | 0.925418 | 0.439881 |
| BABA 1h | 14.7596 | 3.8926 | 1.73315 | 0.451705 |
| Chitin 1h | 7.24413 | 3.18179 | 0.936934 | 0.454185 |
| Epi 1h | 8.12095 | 3.12091 | 1.03666 | 0.487793 |
| SA 1h | 22.045 | 8.36264 | 2.02545 | 1.69524 |
| Me-JA 1h | 25.2821 | 3.5394 | 3.72595 | 0.520487 |
| Control 6h | 8.89877 | 3.35971 | 1.04733 | 0.415896 |
| ABA 6h | 6.95231 | 3.4245 | 0.786057 | 0.392243 |
| ACC 6h | 8.30346 | 3.97853 | 0.851539 | 0.421175 |
| BABA 6h | 7.34781 | 3.74025 | 0.783258 | 0.408333 |
| Chitin 6h | 7.38754 | 3.63298 | 0.821665 | 0.422116 |
| Epi 6h | 12.8935 | 5.7555 | 1.15184 | 0.526363 |
| SA 6h | 7.31473 | 3.48708 | 0.871518 | 0.439987 |
| Me-JA 6h | 22.1281 | 4.32349 | 2.59462 | 0.448036 |
Source Transcript PGSC0003DMT400034695 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G65930.1 | +3 | 4e-36 | 87 | 41/50 (82%) | cytosolic NADP+-dependent isocitrate dehydrogenase | chr1:24539088-24541861 FORWARD LENGTH=410 |
