Probe CUST_25291_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25291_PI426222305 | JHI_St_60k_v1 | DMT400034697 | TTTCACTAAAAATGGCTTTTCAAAAGATCAAAGTGGCTAACCCCATAGTTGAAATGGACG |
All Microarray Probes Designed to Gene DMG400013332
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25432_PI426222305 | JHI_St_60k_v1 | DMT400034698 | AAAGAAATGGCCACTCTATCTTAGCACAAAAAATACCATCCTTAAGAAATACGACGGAAG |
| CUST_25460_PI426222305 | JHI_St_60k_v1 | DMT400034694 | TGAAAGCGCTGAGGCTACTTTGAAATGACCCGTGTCATTTGGAAATTAATTAAGGATAAG |
| CUST_25291_PI426222305 | JHI_St_60k_v1 | DMT400034697 | TTTCACTAAAAATGGCTTTTCAAAAGATCAAAGTGGCTAACCCCATAGTTGAAATGGACG |
| CUST_25361_PI426222305 | JHI_St_60k_v1 | DMT400034696 | TAGCACTTATTATCCACGGATCCAAGATTTGGATCGCTTGGATTGATGACATCGATCCTG |
| CUST_25375_PI426222305 | JHI_St_60k_v1 | DMT400034695 | AGACAGCGTGTCTAACGGCCTTTTGTTATAATCATTATAGTTCTTTCTTTGACTCTTCTT |
Microarray Signals from CUST_25291_PI426222305
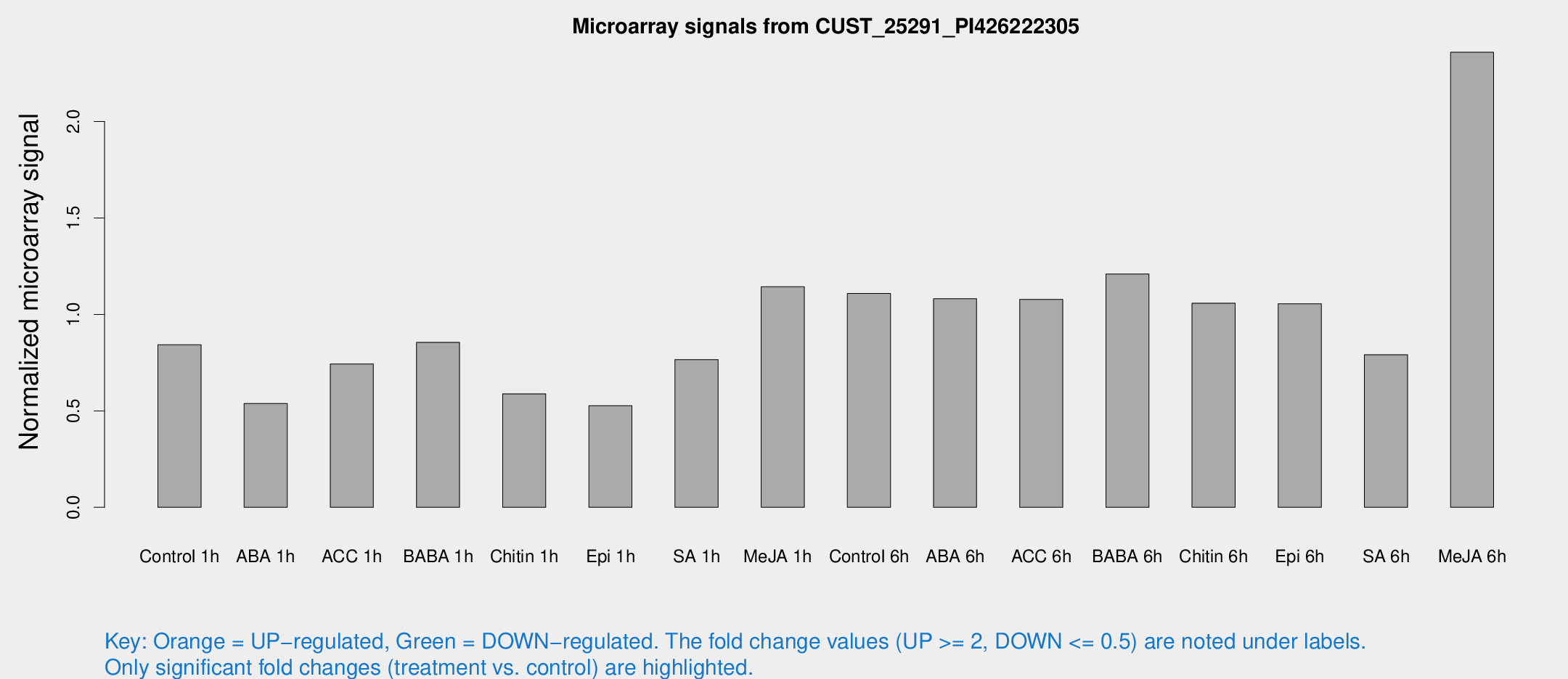
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 18.6244 | 6.29177 | 0.842726 | 0.28583 |
| ABA 1h | 13.3392 | 8.07284 | 0.538254 | 0.340121 |
| ACC 1h | 17.7424 | 5.84694 | 0.743134 | 0.309991 |
| BABA 1h | 17.1856 | 3.80801 | 0.85542 | 0.197555 |
| Chitin 1h | 11.793 | 3.98911 | 0.588116 | 0.217955 |
| Epi 1h | 9.28771 | 3.13582 | 0.526942 | 0.190403 |
| SA 1h | 15.6347 | 3.21637 | 0.766177 | 0.162319 |
| Me-JA 1h | 18.8346 | 3.36465 | 1.14332 | 0.212765 |
| Control 6h | 24.604 | 7.65658 | 1.10894 | 0.315678 |
| ABA 6h | 24.1546 | 6.22563 | 1.08137 | 0.228879 |
| ACC 6h | 27.2227 | 8.14358 | 1.07803 | 0.440117 |
| BABA 6h | 27.0361 | 3.98663 | 1.20991 | 0.185363 |
| Chitin 6h | 22.4893 | 3.79584 | 1.05864 | 0.185026 |
| Epi 6h | 26.4498 | 7.9383 | 1.05529 | 0.360369 |
| SA 6h | 21.2096 | 11.8912 | 0.790954 | 0.459223 |
| Me-JA 6h | 49.3851 | 11.5999 | 2.35967 | 0.505651 |
Source Transcript PGSC0003DMT400034697 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G65930.1 | +2 | 0.0 | 731 | 351/410 (86%) | cytosolic NADP+-dependent isocitrate dehydrogenase | chr1:24539088-24541861 FORWARD LENGTH=410 |
