Probe CUST_24936_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24936_PI426222305 | JHI_St_60k_v1 | DMT400045216 | CTACTGGAAATGCTCGTTTTGGTGAAGTTTCAGAGTCATACACAGTAGATATCAATAAAT |
All Microarray Probes Designed to Gene DMG400017542
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24855_PI426222305 | JHI_St_60k_v1 | DMT400045221 | CCTTACTTGTGCGATGATAGTGAATTTTATTTTTGATGAAAATGGATAGCTATGGCATGC |
| CUST_24920_PI426222305 | JHI_St_60k_v1 | DMT400045220 | GATCCCCCATGTGATAAGATTTTGCAAATTGTAGTTGTTGCTTTTAAGTGCAAATCTCTT |
| CUST_24936_PI426222305 | JHI_St_60k_v1 | DMT400045216 | CTACTGGAAATGCTCGTTTTGGTGAAGTTTCAGAGTCATACACAGTAGATATCAATAAAT |
| CUST_24854_PI426222305 | JHI_St_60k_v1 | DMT400045219 | GATCCCCCATGTGATAAGATTTTGCAAATTGTAGTTGTTGCTTTTAAGTGCAAATCTCTT |
| CUST_24930_PI426222305 | JHI_St_60k_v1 | DMT400045218 | GATCCCCCATGTGATAAGATTTTGCAAATTGTAGTTGTTGCTTTTAAGTGCAAATCTCTT |
Microarray Signals from CUST_24936_PI426222305
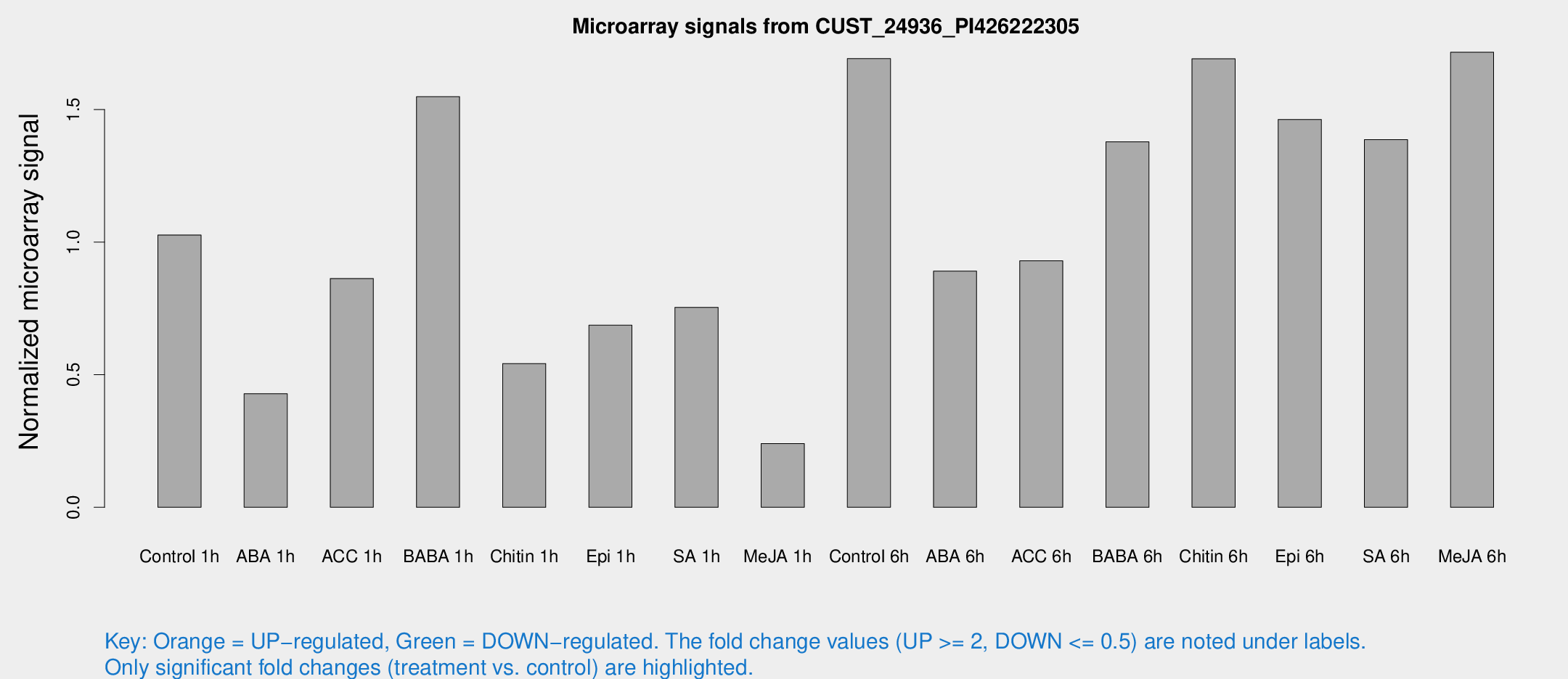
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 47.0831 | 5.50075 | 1.02715 | 0.144523 |
| ABA 1h | 17.3937 | 3.53212 | 0.428638 | 0.0912038 |
| ACC 1h | 43.3791 | 10.8856 | 0.86302 | 0.19742 |
| BABA 1h | 122.235 | 90.4427 | 1.54888 | 1.66397 |
| Chitin 1h | 23.2002 | 5.58335 | 0.542106 | 0.169706 |
| Epi 1h | 30.5934 | 9.21605 | 0.686763 | 0.287963 |
| SA 1h | 37.0061 | 8.15365 | 0.754097 | 0.248329 |
| Me-JA 1h | 9.30163 | 3.6416 | 0.240409 | 0.104173 |
| Control 6h | 86.1982 | 24.9399 | 1.69232 | 0.494477 |
| ABA 6h | 46.5461 | 12.8643 | 0.890828 | 0.227323 |
| ACC 6h | 49.2884 | 7.55472 | 0.92973 | 0.220371 |
| BABA 6h | 70.8184 | 8.76911 | 1.37846 | 0.123003 |
| Chitin 6h | 81.6586 | 6.14065 | 1.69145 | 0.127276 |
| Epi 6h | 79.4079 | 19.2642 | 1.46289 | 0.476901 |
| SA 6h | 68.3128 | 21.673 | 1.38668 | 0.466578 |
| Me-JA 6h | 84.9408 | 22.5993 | 1.71634 | 0.461924 |
Source Transcript PGSC0003DMT400045216 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23910.1 | +1 | 4e-68 | 225 | 131/285 (46%) | NAD(P)-binding Rossmann-fold superfamily protein | chr2:10177902-10179789 FORWARD LENGTH=304 |
