Probe CUST_24854_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24854_PI426222305 | JHI_St_60k_v1 | DMT400045219 | GATCCCCCATGTGATAAGATTTTGCAAATTGTAGTTGTTGCTTTTAAGTGCAAATCTCTT |
All Microarray Probes Designed to Gene DMG400017542
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24855_PI426222305 | JHI_St_60k_v1 | DMT400045221 | CCTTACTTGTGCGATGATAGTGAATTTTATTTTTGATGAAAATGGATAGCTATGGCATGC |
| CUST_24920_PI426222305 | JHI_St_60k_v1 | DMT400045220 | GATCCCCCATGTGATAAGATTTTGCAAATTGTAGTTGTTGCTTTTAAGTGCAAATCTCTT |
| CUST_24936_PI426222305 | JHI_St_60k_v1 | DMT400045216 | CTACTGGAAATGCTCGTTTTGGTGAAGTTTCAGAGTCATACACAGTAGATATCAATAAAT |
| CUST_24854_PI426222305 | JHI_St_60k_v1 | DMT400045219 | GATCCCCCATGTGATAAGATTTTGCAAATTGTAGTTGTTGCTTTTAAGTGCAAATCTCTT |
| CUST_24930_PI426222305 | JHI_St_60k_v1 | DMT400045218 | GATCCCCCATGTGATAAGATTTTGCAAATTGTAGTTGTTGCTTTTAAGTGCAAATCTCTT |
Microarray Signals from CUST_24854_PI426222305
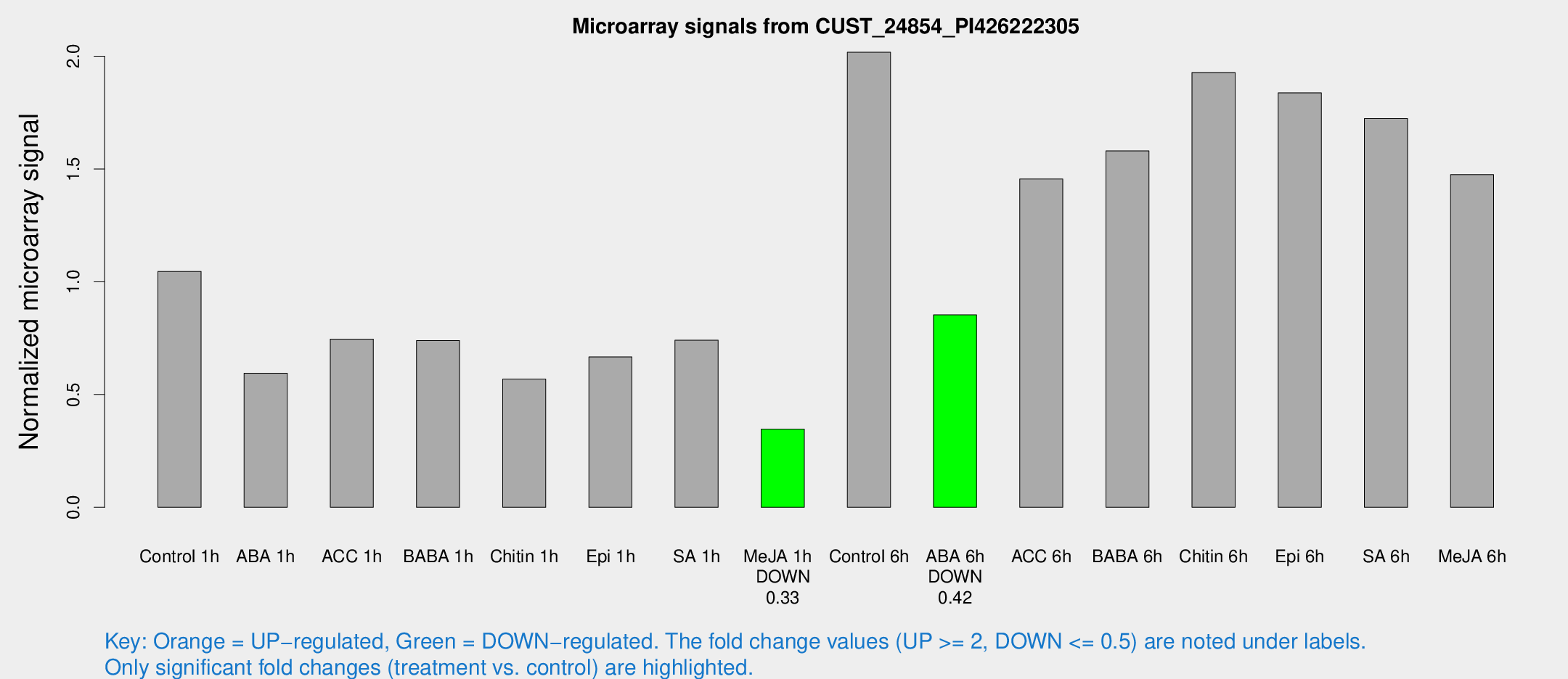
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 612.838 | 71.5535 | 1.04588 | 0.0753882 |
| ABA 1h | 308.69 | 36.101 | 0.594511 | 0.035973 |
| ACC 1h | 509.52 | 158.616 | 0.74599 | 0.263842 |
| BABA 1h | 429.497 | 80.0252 | 0.739002 | 0.0787305 |
| Chitin 1h | 298.603 | 29.8272 | 0.568656 | 0.0336035 |
| Epi 1h | 340.425 | 46.9686 | 0.666537 | 0.0929055 |
| SA 1h | 440.942 | 25.745 | 0.741155 | 0.0563383 |
| Me-JA 1h | 164.501 | 11.9527 | 0.346524 | 0.0215423 |
| Control 6h | 1232.02 | 253.618 | 2.01757 | 0.297596 |
| ABA 6h | 529.814 | 52.4181 | 0.852991 | 0.0496779 |
| ACC 6h | 984.825 | 136.82 | 1.45563 | 0.0843265 |
| BABA 6h | 1046.78 | 149.716 | 1.57992 | 0.192262 |
| Chitin 6h | 1191.33 | 71.9264 | 1.92776 | 0.11151 |
| Epi 6h | 1217.39 | 155.104 | 1.83761 | 0.294752 |
| SA 6h | 1002.38 | 122.32 | 1.72352 | 0.0997892 |
| Me-JA 6h | 923.8 | 259.9 | 1.4746 | 0.318326 |
Source Transcript PGSC0003DMT400045219 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23910.1 | +2 | 6e-74 | 237 | 136/300 (45%) | NAD(P)-binding Rossmann-fold superfamily protein | chr2:10177902-10179789 FORWARD LENGTH=304 |
