Probe CUST_24334_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24334_PI426222305 | JHI_St_60k_v1 | DMT400036081 | GAGAGAGCTGCCACCATGGAAAAGATTAATGATGCTTGTGAAAATTGGGGCTTCTTTGAG |
All Microarray Probes Designed to Gene DMG400013894
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24334_PI426222305 | JHI_St_60k_v1 | DMT400036081 | GAGAGAGCTGCCACCATGGAAAAGATTAATGATGCTTGTGAAAATTGGGGCTTCTTTGAG |
| CUST_24377_PI426222305 | JHI_St_60k_v1 | DMT400036083 | TAGCTAAGTTAAATCCTTTAAACAGGTAATCACGAATGGGAAGTATAAGAGTGTGATGCA |
Microarray Signals from CUST_24334_PI426222305
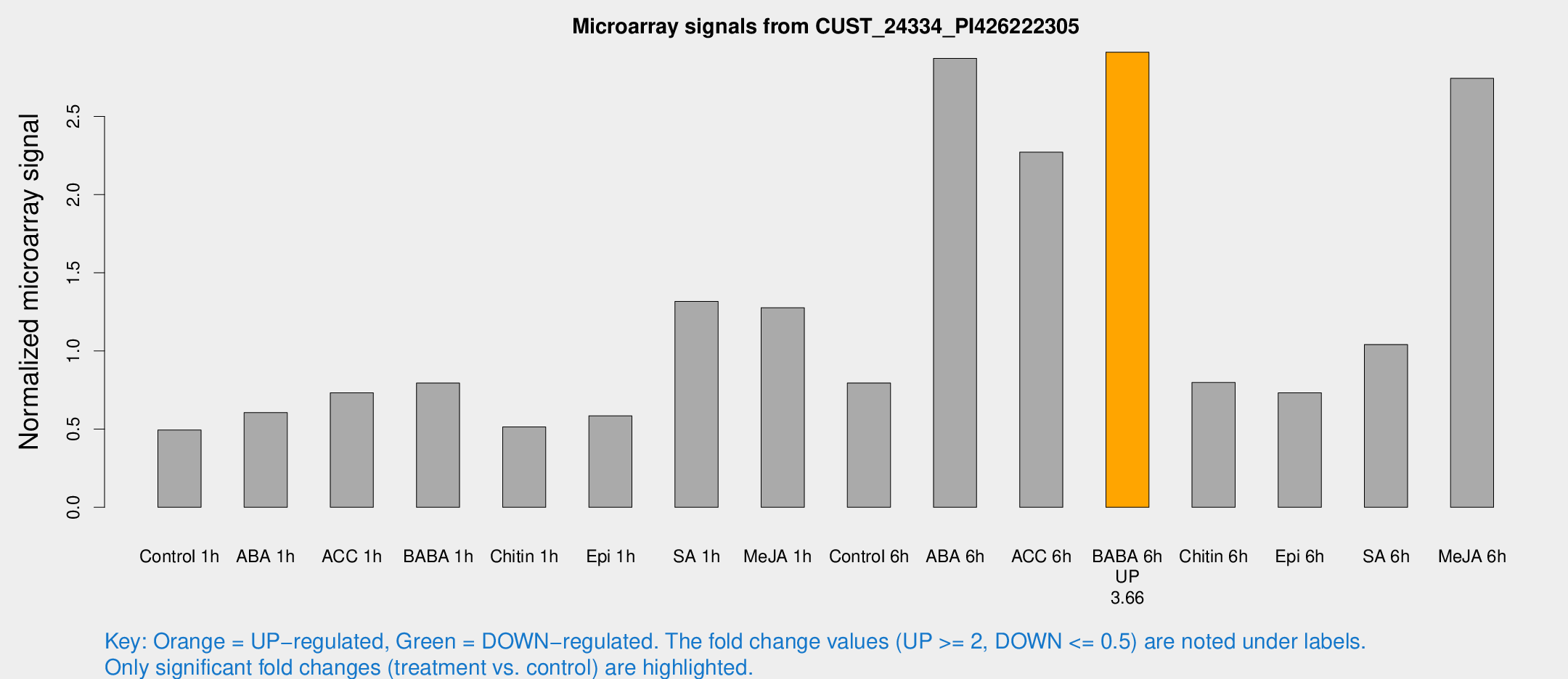
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 154.016 | 50.17 | 0.494124 | 0.1255 |
| ABA 1h | 162.953 | 45.5751 | 0.60592 | 0.165337 |
| ACC 1h | 262.986 | 118.075 | 0.732362 | 0.34268 |
| BABA 1h | 235.978 | 61.0218 | 0.794949 | 0.170444 |
| Chitin 1h | 150.238 | 56.0949 | 0.514368 | 0.186017 |
| Epi 1h | 145.851 | 19.1209 | 0.585324 | 0.0753375 |
| SA 1h | 415.033 | 117.523 | 1.31674 | 0.312246 |
| Me-JA 1h | 307.266 | 62.4809 | 1.27698 | 0.156559 |
| Control 6h | 259.629 | 82.9347 | 0.795143 | 0.26741 |
| ABA 6h | 1029.55 | 404.216 | 2.87182 | 1.22656 |
| ACC 6h | 736.21 | 42.7473 | 2.27153 | 0.353882 |
| BABA 6h | 1020.76 | 344.404 | 2.91106 | 0.891223 |
| Chitin 6h | 248.069 | 44.8976 | 0.798291 | 0.164294 |
| Epi 6h | 241.234 | 42.803 | 0.732916 | 0.107699 |
| SA 6h | 320.188 | 88.6584 | 1.04105 | 0.229962 |
| Me-JA 6h | 786.269 | 111.156 | 2.74377 | 0.192998 |
Source Transcript PGSC0003DMT400036081 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05010.1 | +2 | 8e-160 | 459 | 235/311 (76%) | ethylene-forming enzyme | chr1:1431419-1432695 REVERSE LENGTH=323 |
