Probe CUST_23023_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23023_PI426222305 | JHI_St_60k_v1 | DMT400060913 | TGGTGATCCTGAACGTGCCCTAGTTATGGCTTTTAAAGCTATTGCTAATATGGCTGATAG |
All Microarray Probes Designed to Gene DMG400023693
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22876_PI426222305 | JHI_St_60k_v1 | DMT400060916 | GGTATTGGGACAAATTTGCTCCTTCATTACGTAAGAACAAGTCTTACCCGTGTCTTTTTT |
| CUST_22935_PI426222305 | JHI_St_60k_v1 | DMT400060912 | GGCTTTTAAAGCTATTGCTAATATGGCTGATAGGTAAAACTTATCGAACTTCAACCAGTG |
| CUST_22980_PI426222305 | JHI_St_60k_v1 | DMT400060914 | TGGTGATCCTGAACGTGCCCTAGTTATGGCTTTTAAAGCTATTGCTAATATGGCTGATAG |
| CUST_23023_PI426222305 | JHI_St_60k_v1 | DMT400060913 | TGGTGATCCTGAACGTGCCCTAGTTATGGCTTTTAAAGCTATTGCTAATATGGCTGATAG |
| CUST_23071_PI426222305 | JHI_St_60k_v1 | DMT400060915 | GAATGCAATTATTTTTGTTGGATTAGTCAAGTGTGTCCCCCATTGTTGTGTGCATTAATT |
Microarray Signals from CUST_23023_PI426222305
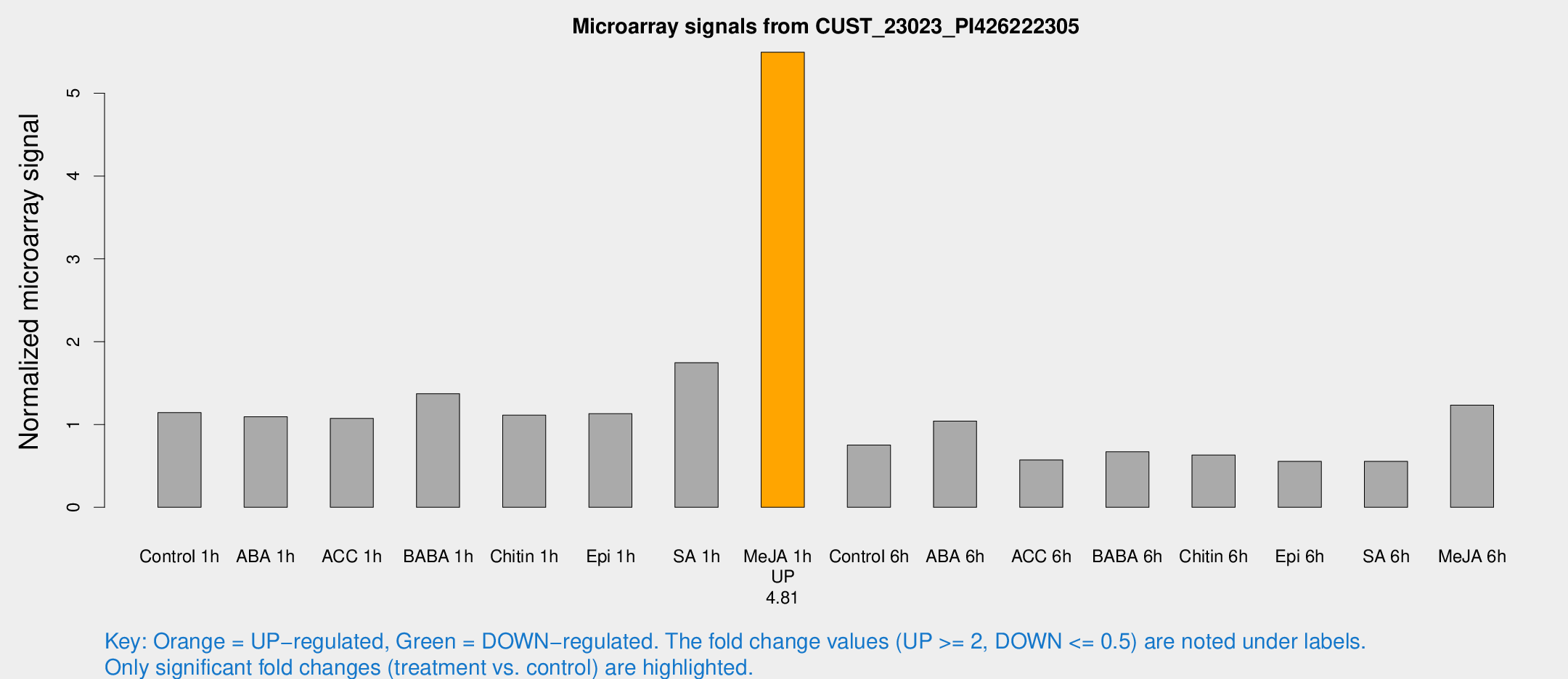
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 360.422 | 68.6537 | 1.14289 | 0.156674 |
| ABA 1h | 340.925 | 128.46 | 1.09377 | 0.366952 |
| ACC 1h | 367.976 | 101.02 | 1.07295 | 0.274328 |
| BABA 1h | 442.585 | 123.345 | 1.37182 | 0.311556 |
| Chitin 1h | 306.745 | 23.5314 | 1.11245 | 0.065918 |
| Epi 1h | 300.391 | 24.2282 | 1.13063 | 0.0877188 |
| SA 1h | 562.074 | 91.9476 | 1.7457 | 0.250725 |
| Me-JA 1h | 1376.63 | 107.599 | 5.49557 | 0.31755 |
| Control 6h | 260.431 | 79.2966 | 0.752078 | 0.232343 |
| ABA 6h | 349.268 | 59.0523 | 1.04207 | 0.156228 |
| ACC 6h | 204.199 | 30.2734 | 0.571388 | 0.0442609 |
| BABA 6h | 232.674 | 29.5452 | 0.669895 | 0.0691973 |
| Chitin 6h | 207.844 | 23.4185 | 0.631873 | 0.0698983 |
| Epi 6h | 202.533 | 52.7677 | 0.554024 | 0.140828 |
| SA 6h | 175.858 | 37.2536 | 0.554609 | 0.0677082 |
| Me-JA 6h | 388.242 | 67.9593 | 1.23442 | 0.163784 |
Source Transcript PGSC0003DMT400060913 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G10330.1 | +1 | 1e-160 | 462 | 238/313 (76%) | Cyclin-like family protein | chr3:3199907-3201642 FORWARD LENGTH=312 |
