Probe CUST_22980_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22980_PI426222305 | JHI_St_60k_v1 | DMT400060914 | TGGTGATCCTGAACGTGCCCTAGTTATGGCTTTTAAAGCTATTGCTAATATGGCTGATAG |
All Microarray Probes Designed to Gene DMG400023693
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22876_PI426222305 | JHI_St_60k_v1 | DMT400060916 | GGTATTGGGACAAATTTGCTCCTTCATTACGTAAGAACAAGTCTTACCCGTGTCTTTTTT |
| CUST_22935_PI426222305 | JHI_St_60k_v1 | DMT400060912 | GGCTTTTAAAGCTATTGCTAATATGGCTGATAGGTAAAACTTATCGAACTTCAACCAGTG |
| CUST_22980_PI426222305 | JHI_St_60k_v1 | DMT400060914 | TGGTGATCCTGAACGTGCCCTAGTTATGGCTTTTAAAGCTATTGCTAATATGGCTGATAG |
| CUST_23023_PI426222305 | JHI_St_60k_v1 | DMT400060913 | TGGTGATCCTGAACGTGCCCTAGTTATGGCTTTTAAAGCTATTGCTAATATGGCTGATAG |
| CUST_23071_PI426222305 | JHI_St_60k_v1 | DMT400060915 | GAATGCAATTATTTTTGTTGGATTAGTCAAGTGTGTCCCCCATTGTTGTGTGCATTAATT |
Microarray Signals from CUST_22980_PI426222305
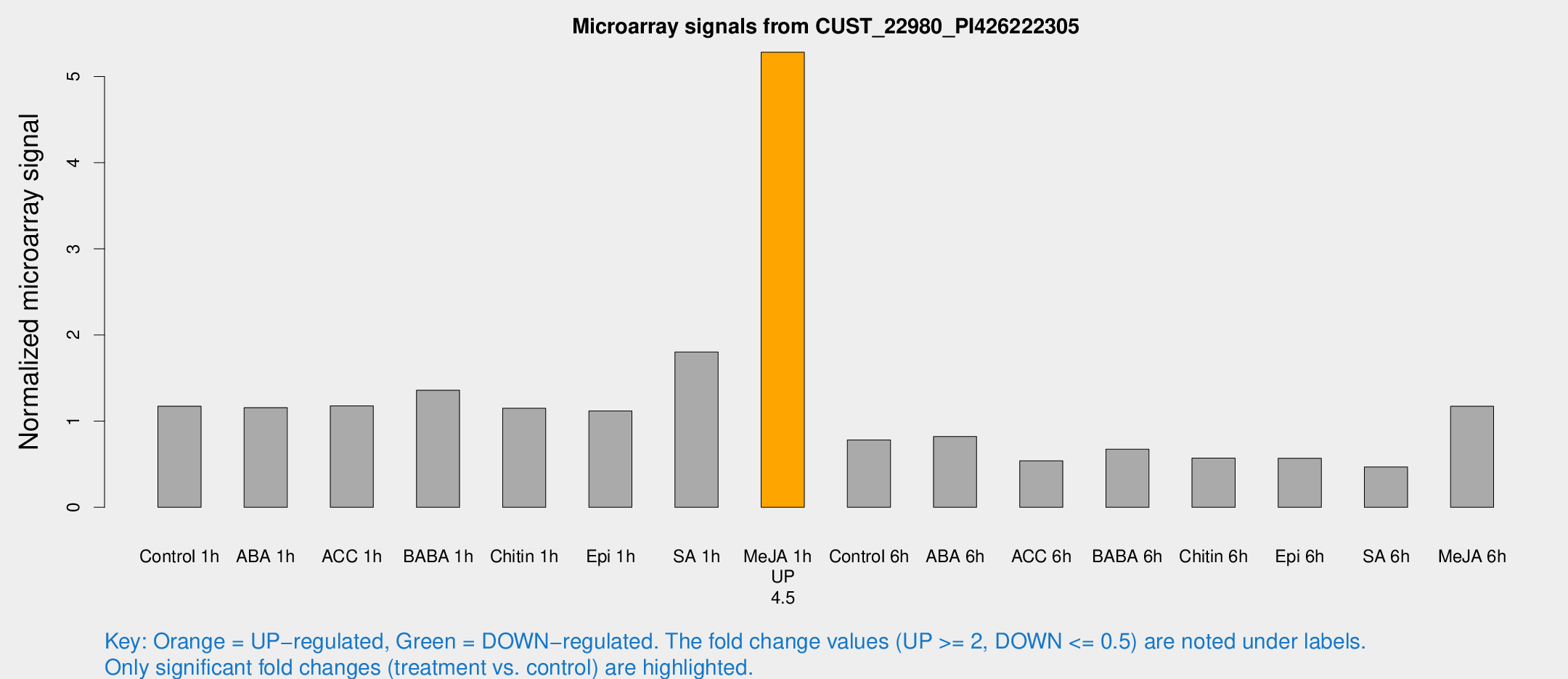
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 388.985 | 72.8042 | 1.17388 | 0.161349 |
| ABA 1h | 370.608 | 129.347 | 1.15705 | 0.340727 |
| ACC 1h | 423.796 | 121.621 | 1.17759 | 0.29679 |
| BABA 1h | 445.255 | 99.9045 | 1.35871 | 0.207169 |
| Chitin 1h | 332.687 | 20.9417 | 1.14932 | 0.067213 |
| Epi 1h | 313.506 | 29.7714 | 1.11898 | 0.0862787 |
| SA 1h | 610.749 | 103.205 | 1.80168 | 0.272144 |
| Me-JA 1h | 1392.41 | 111.696 | 5.28238 | 0.305216 |
| Control 6h | 281.554 | 80.576 | 0.782124 | 0.222864 |
| ABA 6h | 292.656 | 61.2743 | 0.821873 | 0.163826 |
| ACC 6h | 205.898 | 38.2997 | 0.540581 | 0.0610241 |
| BABA 6h | 244.856 | 26.9043 | 0.674301 | 0.0656548 |
| Chitin 6h | 195.358 | 11.8106 | 0.572198 | 0.0345866 |
| Epi 6h | 214.64 | 46.0331 | 0.569237 | 0.120245 |
| SA 6h | 168.298 | 50.7416 | 0.469012 | 0.127563 |
| Me-JA 6h | 388.865 | 70.4949 | 1.17249 | 0.141009 |
Source Transcript PGSC0003DMT400060914 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G10330.1 | +1 | 6e-133 | 394 | 206/273 (75%) | Cyclin-like family protein | chr3:3199907-3201642 FORWARD LENGTH=312 |
