Probe CUST_23014_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23014_PI426222305 | JHI_St_60k_v1 | DMT400021132 | ATTAGCGAAGTGGTTCCCATGTGGAGAGTAACTTACAAGACATGGTTGTGCCACCTATCT |
All Microarray Probes Designed to Gene DMG400008183
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22942_PI426222305 | JHI_St_60k_v1 | DMT400021130 | GCGACTTCCAATCCACTAGTCAATCAGAAATCAACAAAGAGAAAGGAACGCGAGAAAAAG |
| CUST_23014_PI426222305 | JHI_St_60k_v1 | DMT400021132 | ATTAGCGAAGTGGTTCCCATGTGGAGAGTAACTTACAAGACATGGTTGTGCCACCTATCT |
| CUST_23088_PI426222305 | JHI_St_60k_v1 | DMT400021129 | CCCATTTAGTGTAGGAGCCAGAATTTTCATTTTCTCCTGTAGGTAACATCTAAAGAAGTA |
Microarray Signals from CUST_23014_PI426222305
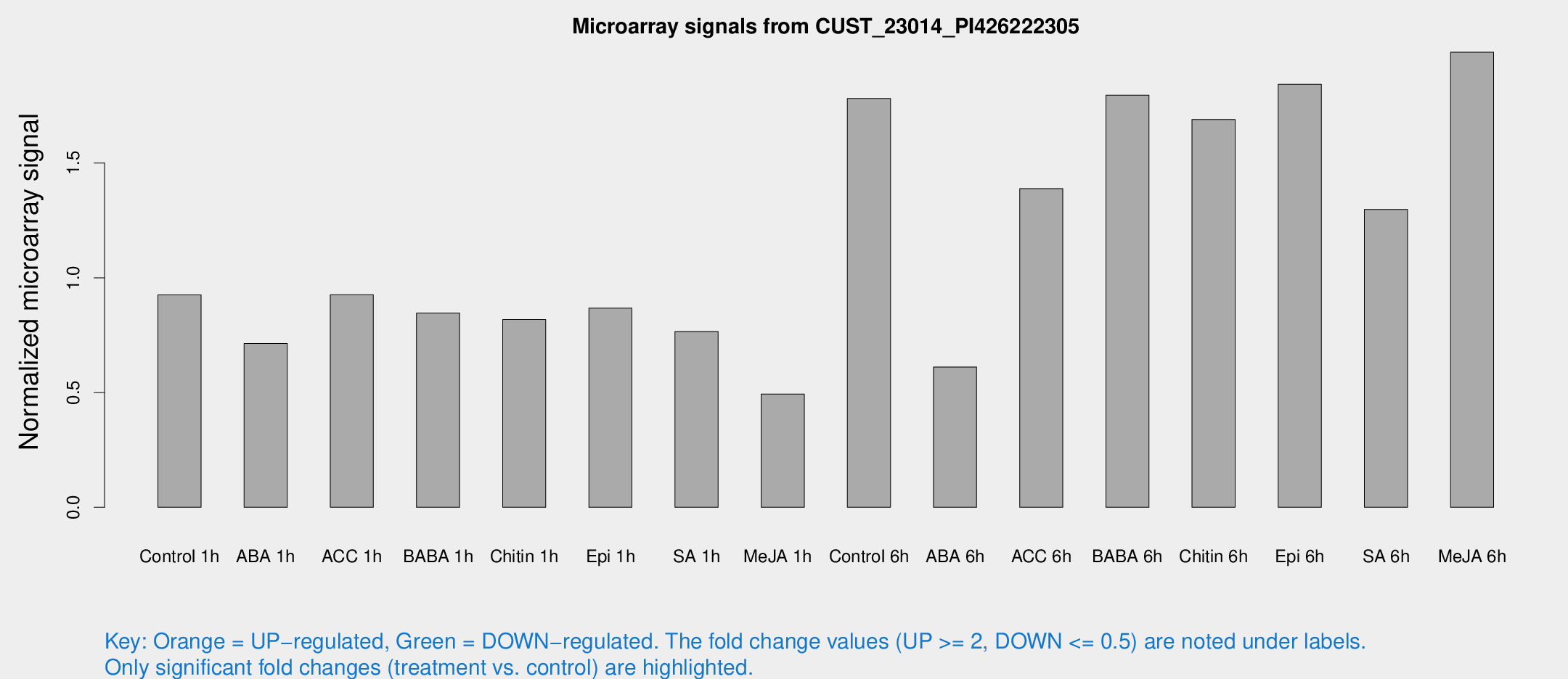
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1273.1 | 186.723 | 0.925872 | 0.0796872 |
| ABA 1h | 890.268 | 186.495 | 0.713705 | 0.136801 |
| ACC 1h | 1322.87 | 239.52 | 0.926053 | 0.109852 |
| BABA 1h | 1152.12 | 242.351 | 0.846675 | 0.111856 |
| Chitin 1h | 998.316 | 83.9163 | 0.818155 | 0.0473321 |
| Epi 1h | 1045.56 | 181.095 | 0.868298 | 0.139509 |
| SA 1h | 1079.63 | 143.342 | 0.766017 | 0.0910355 |
| Me-JA 1h | 546.268 | 43.874 | 0.493393 | 0.0286857 |
| Control 6h | 2807.82 | 934.534 | 1.78139 | 0.652046 |
| ABA 6h | 881.919 | 75.293 | 0.61116 | 0.0353961 |
| ACC 6h | 2260.64 | 523.318 | 1.38888 | 0.129651 |
| BABA 6h | 2783.76 | 433.148 | 1.79535 | 0.246641 |
| Chitin 6h | 2425.41 | 140.147 | 1.68988 | 0.0976123 |
| Epi 6h | 2925.88 | 629.029 | 1.84294 | 0.445152 |
| SA 6h | 1810.66 | 350.457 | 1.29826 | 0.132465 |
| Me-JA 6h | 2810.3 | 591.003 | 1.98302 | 0.350677 |
Source Transcript PGSC0003DMT400021132 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G42300.1 | +2 | 9e-07 | 49 | 28/79 (35%) | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | chr2:17621542-17624635 FORWARD LENGTH=327 |
