Probe CUST_22942_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22942_PI426222305 | JHI_St_60k_v1 | DMT400021130 | GCGACTTCCAATCCACTAGTCAATCAGAAATCAACAAAGAGAAAGGAACGCGAGAAAAAG |
All Microarray Probes Designed to Gene DMG400008183
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22942_PI426222305 | JHI_St_60k_v1 | DMT400021130 | GCGACTTCCAATCCACTAGTCAATCAGAAATCAACAAAGAGAAAGGAACGCGAGAAAAAG |
| CUST_23014_PI426222305 | JHI_St_60k_v1 | DMT400021132 | ATTAGCGAAGTGGTTCCCATGTGGAGAGTAACTTACAAGACATGGTTGTGCCACCTATCT |
| CUST_23088_PI426222305 | JHI_St_60k_v1 | DMT400021129 | CCCATTTAGTGTAGGAGCCAGAATTTTCATTTTCTCCTGTAGGTAACATCTAAAGAAGTA |
Microarray Signals from CUST_22942_PI426222305
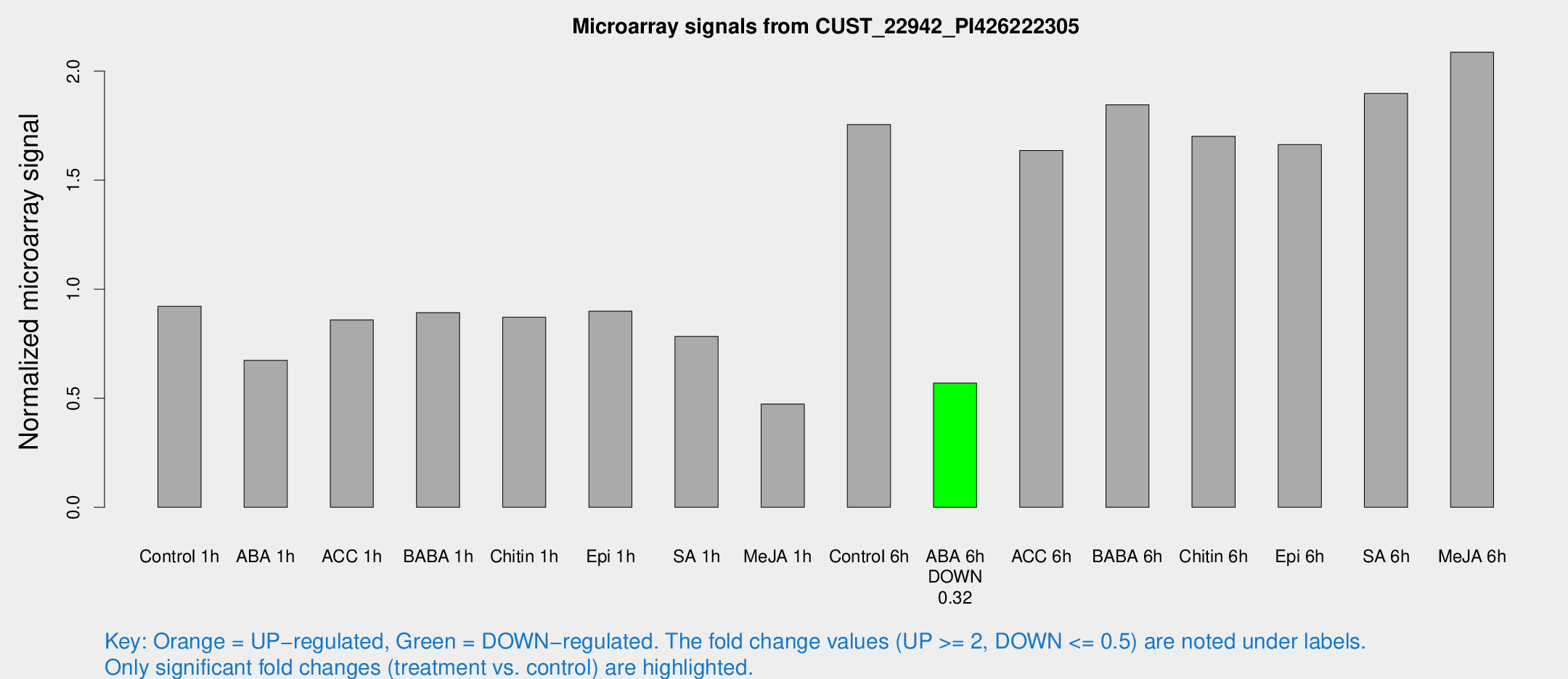
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1309.07 | 144.462 | 0.921598 | 0.053258 |
| ABA 1h | 862.346 | 152.383 | 0.673681 | 0.0929192 |
| ACC 1h | 1283.59 | 232.951 | 0.8591 | 0.100954 |
| BABA 1h | 1268.01 | 284.48 | 0.892308 | 0.134978 |
| Chitin 1h | 1111.32 | 109.385 | 0.871429 | 0.068194 |
| Epi 1h | 1117.28 | 163.188 | 0.899273 | 0.116035 |
| SA 1h | 1135.85 | 76.4196 | 0.783843 | 0.0678271 |
| Me-JA 1h | 544.875 | 34.5692 | 0.473523 | 0.0274894 |
| Control 6h | 2690.92 | 685.049 | 1.75485 | 0.394665 |
| ABA 6h | 860.266 | 95.6666 | 0.569613 | 0.0402784 |
| ACC 6h | 2767.89 | 586.461 | 1.6357 | 0.21456 |
| BABA 6h | 3050.88 | 637.112 | 1.84525 | 0.380701 |
| Chitin 6h | 2591.84 | 357.363 | 1.70086 | 0.270944 |
| Epi 6h | 2780.95 | 646.59 | 1.66365 | 0.476101 |
| SA 6h | 2994.7 | 1035.38 | 1.89761 | 1.07105 |
| Me-JA 6h | 3078.88 | 644.375 | 2.08648 | 0.355258 |
Source Transcript PGSC0003DMT400021130 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G42300.1 | +1 | 5e-63 | 215 | 152/323 (47%) | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | chr2:17621542-17624635 FORWARD LENGTH=327 |
