Probe CUST_2086_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2086_PI426222305 | JHI_St_60k_v1 | DMT400027148 | ACCGTAAGAAGTCCTATAGTAGTTGCTCCAGCGCTTGATGCCCCAAATGAAAATGATTGA |
All Microarray Probes Designed to Gene DMG400010470
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2500_PI426222305 | JHI_St_60k_v1 | DMT400027149 | CAGTAGCAACATTCACACAATAGGACTGAAATACTTAGATGTTATGTGGAAGCTAACATT |
| CUST_2086_PI426222305 | JHI_St_60k_v1 | DMT400027148 | ACCGTAAGAAGTCCTATAGTAGTTGCTCCAGCGCTTGATGCCCCAAATGAAAATGATTGA |
Microarray Signals from CUST_2086_PI426222305
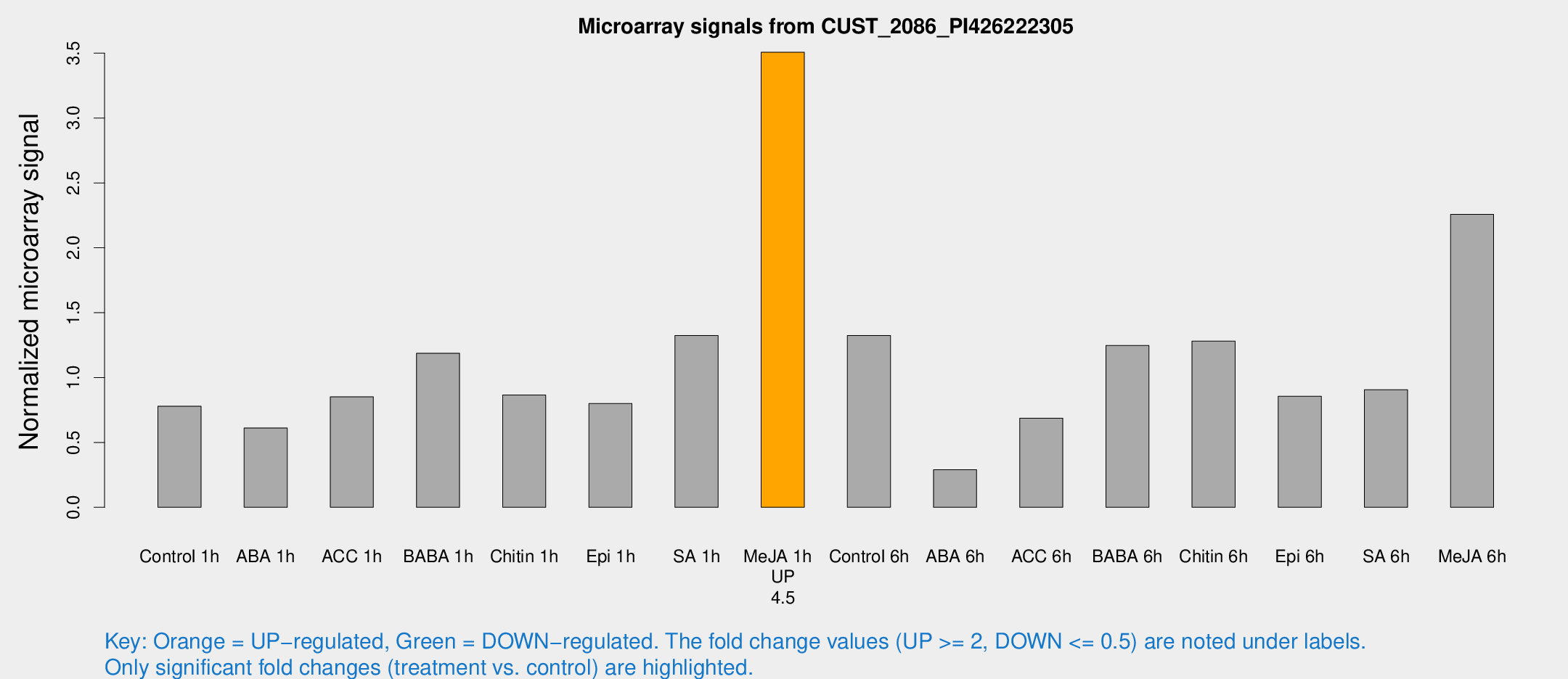
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19341.2 | 3873.72 | 0.779369 | 0.171733 |
| ABA 1h | 14059.8 | 4348.36 | 0.611673 | 0.146471 |
| ACC 1h | 22785.9 | 7386.55 | 0.851215 | 0.24261 |
| BABA 1h | 27496.2 | 3253.65 | 1.18723 | 0.0919665 |
| Chitin 1h | 18512.4 | 1072.28 | 0.86577 | 0.103613 |
| Epi 1h | 17170.5 | 3345.08 | 0.799453 | 0.172424 |
| SA 1h | 36825.5 | 11995.8 | 1.32428 | 0.655369 |
| Me-JA 1h | 68462.4 | 6365.75 | 3.50696 | 0.202475 |
| Control 6h | 34134.3 | 9807.05 | 1.32386 | 0.27857 |
| ABA 6h | 8733.9 | 3814.29 | 0.289678 | 0.154206 |
| ACC 6h | 19652.3 | 4433.31 | 0.686857 | 0.0965128 |
| BABA 6h | 33307.2 | 2623.16 | 1.2471 | 0.119958 |
| Chitin 6h | 33771.2 | 7442.78 | 1.28044 | 0.27913 |
| Epi 6h | 24309.9 | 5333.88 | 0.856396 | 0.299488 |
| SA 6h | 21384.2 | 1755.46 | 0.906212 | 0.0895163 |
| Me-JA 6h | 54067.4 | 6399.07 | 2.25755 | 0.198404 |
Source Transcript PGSC0003DMT400027148 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc10g084320.1 | +1 | 0.0 | 1342 | 665/702 (95%) | evidence_code:10F1H1E1IEG genomic_reference:SL2.50ch10 gene_region:63240593-63242908 transcript_region:SL2.50ch10:63240593..63242908+ go_terms:GO:0006508,GO:0043086 functional_description:Subtilisin-like protease (AHRD V1 **-- O82777_SOLLC); contains Interpro domain(s) IPR015500 Peptidase S8, subtilisin-related |
| TAIR PP10 | AT5G67090.1 | +1 | 2e-176 | 526 | 307/717 (43%) | Subtilisin-like serine endopeptidase family protein | chr5:26774111-26776321 REVERSE LENGTH=736 |
