Probe CUST_2079_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2079_PI426222305 | JHI_St_60k_v1 | DMT400028648 | CCTATCATTTCCGAGTTTAGAAAACATGTATGATTTGAACGAAAACTGCAAGGCGCCTCT |
All Microarray Probes Designed to Gene DMG400011027
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2071_PI426222305 | JHI_St_60k_v1 | DMT400028649 | GGGAACTTTGACATATTGCTTCCTAGTTGGTAGGACAATATTTTCGTGGAAAATAAATGA |
| CUST_2079_PI426222305 | JHI_St_60k_v1 | DMT400028648 | CCTATCATTTCCGAGTTTAGAAAACATGTATGATTTGAACGAAAACTGCAAGGCGCCTCT |
Microarray Signals from CUST_2079_PI426222305
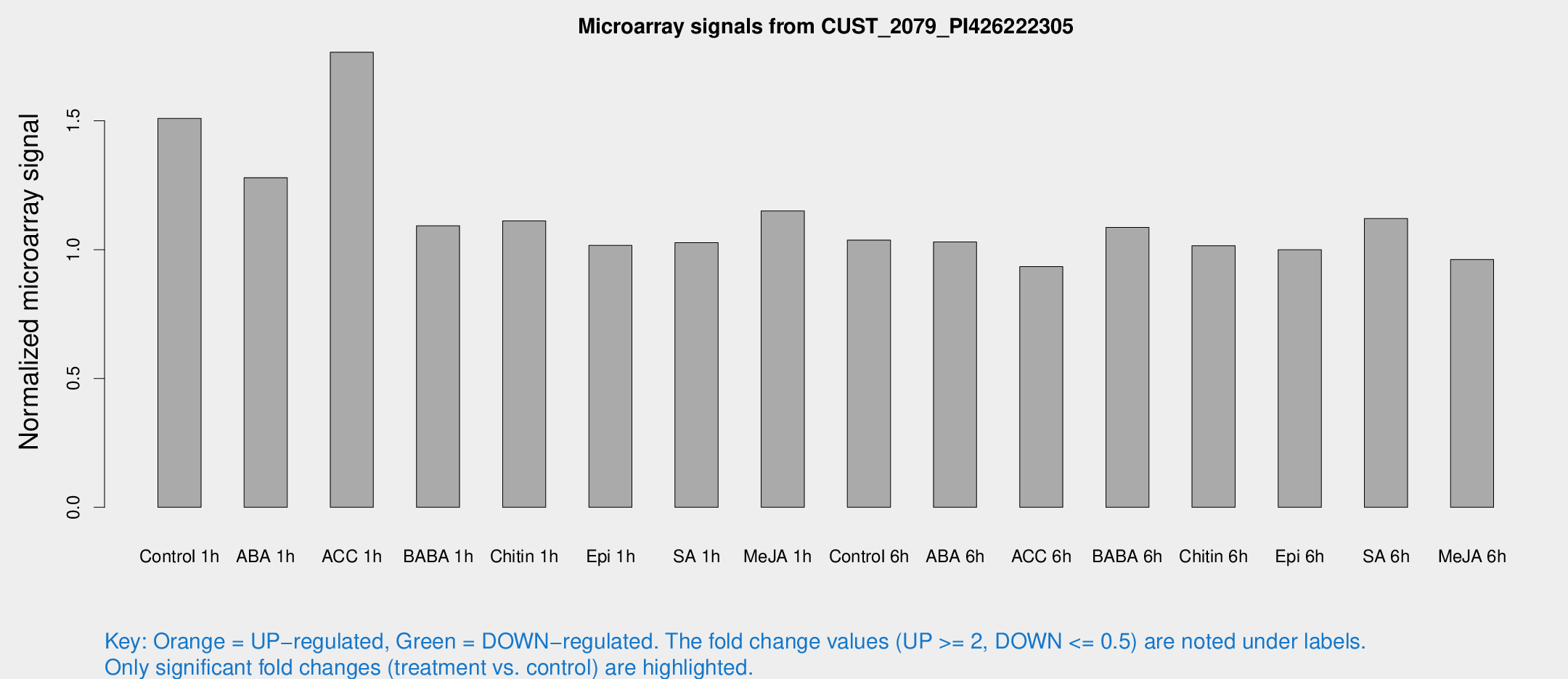
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.1392 | 3.72977 | 1.50929 | 0.590345 |
| ABA 1h | 8.36745 | 3.43033 | 1.27944 | 0.611836 |
| ACC 1h | 12.8975 | 4.54954 | 1.76622 | 0.684988 |
| BABA 1h | 7.40874 | 3.87266 | 1.09273 | 0.584466 |
| Chitin 1h | 7.13123 | 3.63134 | 1.11192 | 0.585734 |
| Epi 1h | 6.09352 | 3.54801 | 1.01629 | 0.589188 |
| SA 1h | 7.40311 | 3.67499 | 1.02729 | 0.529162 |
| Me-JA 1h | 6.50533 | 3.78641 | 1.15015 | 0.666159 |
| Control 6h | 7.22031 | 3.87152 | 1.03728 | 0.562016 |
| ABA 6h | 7.62654 | 3.97402 | 1.02978 | 0.546924 |
| ACC 6h | 7.54393 | 4.46753 | 0.934162 | 0.541536 |
| BABA 6h | 8.52399 | 4.26975 | 1.08656 | 0.560886 |
| Chitin 6h | 7.45522 | 4.26921 | 1.01536 | 0.58102 |
| Epi 6h | 7.92518 | 4.68962 | 0.9994 | 0.579306 |
| SA 6h | 7.68939 | 4.13404 | 1.12077 | 0.608926 |
| Me-JA 6h | 6.60443 | 3.82463 | 0.961786 | 0.556931 |
Source Transcript PGSC0003DMT400028648 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc10g085230.1 | +2 | 0.0 | 871 | 441/476 (93%) | evidence_code:10F1H1E1IEG genomic_reference:SL2.50ch10 gene_region:63824666-63826235 transcript_region:SL2.50ch10:63824666..63826235+ go_terms:GO:0080044 functional_description:UDP-glucosyltransferase (AHRD V1 **** C0LNQ9_MAIZE); contains Interpro domain(s) IPR002213 UDP-glucuronosyl/UDP-glucosyltransferase |
| TAIR PP10 | AT3G11340.1 | +2 | 1e-125 | 380 | 195/444 (44%) | UDP-Glycosyltransferase superfamily protein | chr3:3556728-3558149 FORWARD LENGTH=447 |
