Probe CUST_2071_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2071_PI426222305 | JHI_St_60k_v1 | DMT400028649 | GGGAACTTTGACATATTGCTTCCTAGTTGGTAGGACAATATTTTCGTGGAAAATAAATGA |
All Microarray Probes Designed to Gene DMG400011027
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2071_PI426222305 | JHI_St_60k_v1 | DMT400028649 | GGGAACTTTGACATATTGCTTCCTAGTTGGTAGGACAATATTTTCGTGGAAAATAAATGA |
| CUST_2079_PI426222305 | JHI_St_60k_v1 | DMT400028648 | CCTATCATTTCCGAGTTTAGAAAACATGTATGATTTGAACGAAAACTGCAAGGCGCCTCT |
Microarray Signals from CUST_2071_PI426222305
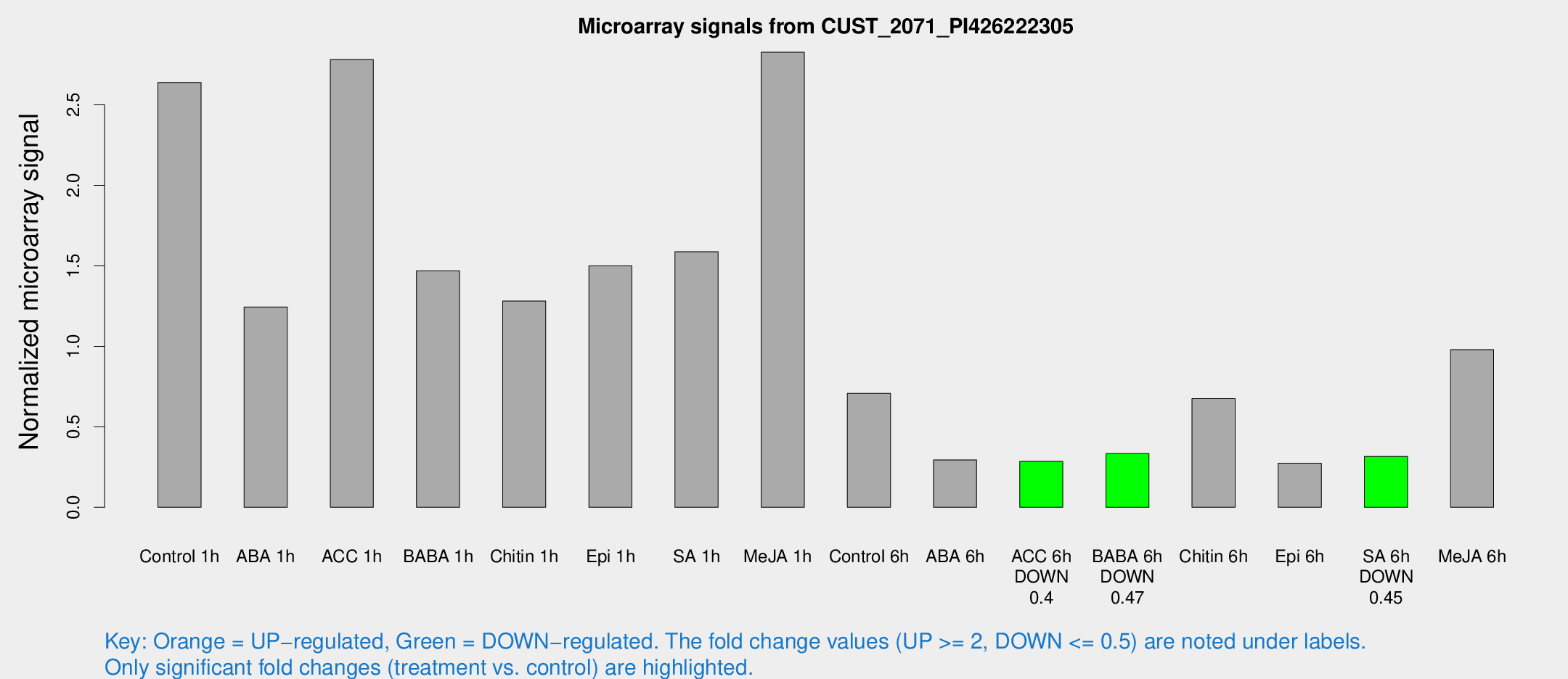
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 503.761 | 58.9336 | 2.63825 | 0.195631 |
| ABA 1h | 211.367 | 27.5614 | 1.24389 | 0.119093 |
| ACC 1h | 587.244 | 154.264 | 2.78085 | 0.687343 |
| BABA 1h | 273.586 | 41.5177 | 1.46897 | 0.102742 |
| Chitin 1h | 223.385 | 39.278 | 1.28074 | 0.228006 |
| Epi 1h | 252.593 | 44.5882 | 1.4998 | 0.270934 |
| SA 1h | 352.392 | 137.238 | 1.58699 | 0.628478 |
| Me-JA 1h | 440.206 | 48.5591 | 2.82644 | 0.303974 |
| Control 6h | 136.752 | 21.5488 | 0.70725 | 0.0710398 |
| ABA 6h | 71.4042 | 32.4238 | 0.294286 | 0.162833 |
| ACC 6h | 63.0371 | 9.81264 | 0.284818 | 0.0250607 |
| BABA 6h | 70.9367 | 6.06959 | 0.33361 | 0.0399397 |
| Chitin 6h | 137.582 | 16.5726 | 0.674956 | 0.10626 |
| Epi 6h | 64.5666 | 18.9446 | 0.274289 | 0.127319 |
| SA 6h | 60.0553 | 8.52287 | 0.31606 | 0.0469867 |
| Me-JA 6h | 201.728 | 64.6363 | 0.97916 | 0.246414 |
Source Transcript PGSC0003DMT400028649 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc10g085230.1 | +2 | 0.0 | 577 | 298/322 (93%) | evidence_code:10F1H1E1IEG genomic_reference:SL2.50ch10 gene_region:63824666-63826235 transcript_region:SL2.50ch10:63824666..63826235+ go_terms:GO:0080044 functional_description:UDP-glucosyltransferase (AHRD V1 **** C0LNQ9_MAIZE); contains Interpro domain(s) IPR002213 UDP-glucuronosyl/UDP-glucosyltransferase |
| TAIR PP10 | AT5G59590.1 | +2 | 5e-94 | 293 | 141/281 (50%) | UDP-glucosyl transferase 76E2 | chr5:24009152-24010585 REVERSE LENGTH=449 |
