Probe CUST_20530_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20530_PI426222305 | JHI_St_60k_v1 | DMT400021304 | ATGAAATAGATAGTGAGAAGAAATGATCCGAATATTTGTCACAGTGTGAGCTTTTCCAGT |
All Microarray Probes Designed to Gene DMG400008259
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20464_PI426222305 | JHI_St_60k_v1 | DMT400021305 | GTCAGCACGTTTGTCCCATCAAAGACTACTGCACTATATAATATTGATCGACTCACTAAC |
| CUST_20520_PI426222305 | JHI_St_60k_v1 | DMT400021303 | TTCAGTCTGACCATTTCTATAAATGATCCGAATATTTGTCACAGTGTGAGCTTTTCCAGT |
| CUST_20530_PI426222305 | JHI_St_60k_v1 | DMT400021304 | ATGAAATAGATAGTGAGAAGAAATGATCCGAATATTTGTCACAGTGTGAGCTTTTCCAGT |
| CUST_20540_PI426222305 | JHI_St_60k_v1 | DMT400021306 | ATGAAATAGATAGTGAGAAGAAATGATCCGAATATTTGTCACAGTGTGAGCTTTTCCAGT |
Microarray Signals from CUST_20530_PI426222305
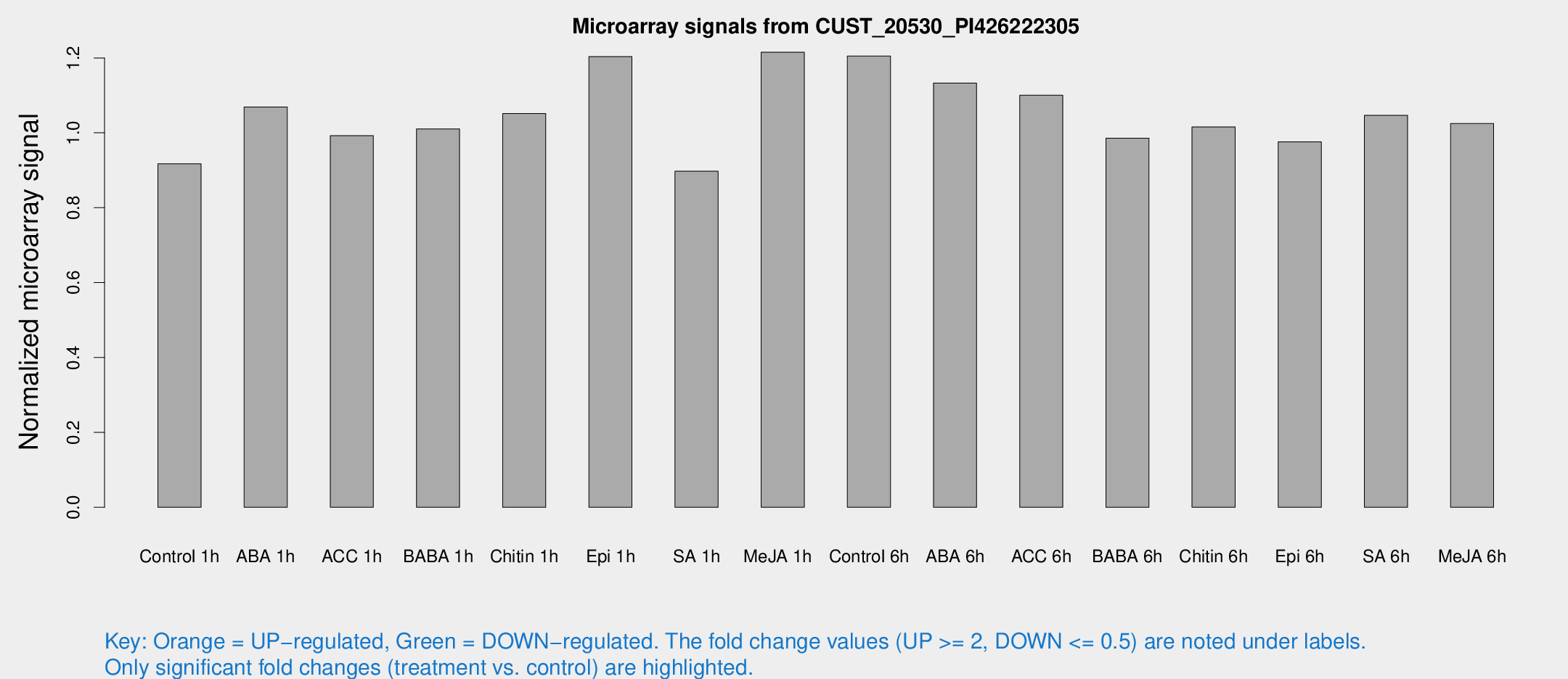
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.44117 | 3.15505 | 0.917188 | 0.531195 |
| ABA 1h | 5.61317 | 3.10744 | 1.06902 | 0.593333 |
| ACC 1h | 6.04293 | 3.48138 | 0.992419 | 0.571689 |
| BABA 1h | 5.78636 | 3.35947 | 1.01053 | 0.585155 |
| Chitin 1h | 5.57895 | 3.25507 | 1.05156 | 0.603391 |
| Epi 1h | 6.21625 | 3.16186 | 1.20349 | 0.625072 |
| SA 1h | 5.46924 | 3.17107 | 0.897441 | 0.519694 |
| Me-JA 1h | 5.89554 | 3.31064 | 1.21527 | 0.679257 |
| Control 6h | 7.36009 | 3.52322 | 1.20507 | 0.610842 |
| ABA 6h | 7.30844 | 3.5039 | 1.13283 | 0.573027 |
| ACC 6h | 7.57815 | 4.06111 | 1.10056 | 0.579849 |
| BABA 6h | 6.56174 | 3.82218 | 0.9859 | 0.572098 |
| Chitin 6h | 6.40928 | 3.71398 | 1.01567 | 0.588277 |
| Epi 6h | 6.59257 | 3.86227 | 0.976145 | 0.565652 |
| SA 6h | 6.13482 | 3.553 | 1.04658 | 0.60606 |
| Me-JA 6h | 6.05131 | 3.4311 | 1.02492 | 0.58055 |
Source Transcript PGSC0003DMT400021304 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G18200.1 | +3 | 6e-103 | 214 | 124/185 (67%) | UTP:galactose-1-phosphate uridylyltransferases;ribose-5-phosphate adenylyltransferases | chr5:6015270-6016555 FORWARD LENGTH=351 |
