Probe CUST_20464_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20464_PI426222305 | JHI_St_60k_v1 | DMT400021305 | GTCAGCACGTTTGTCCCATCAAAGACTACTGCACTATATAATATTGATCGACTCACTAAC |
All Microarray Probes Designed to Gene DMG400008259
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20464_PI426222305 | JHI_St_60k_v1 | DMT400021305 | GTCAGCACGTTTGTCCCATCAAAGACTACTGCACTATATAATATTGATCGACTCACTAAC |
| CUST_20520_PI426222305 | JHI_St_60k_v1 | DMT400021303 | TTCAGTCTGACCATTTCTATAAATGATCCGAATATTTGTCACAGTGTGAGCTTTTCCAGT |
| CUST_20530_PI426222305 | JHI_St_60k_v1 | DMT400021304 | ATGAAATAGATAGTGAGAAGAAATGATCCGAATATTTGTCACAGTGTGAGCTTTTCCAGT |
| CUST_20540_PI426222305 | JHI_St_60k_v1 | DMT400021306 | ATGAAATAGATAGTGAGAAGAAATGATCCGAATATTTGTCACAGTGTGAGCTTTTCCAGT |
Microarray Signals from CUST_20464_PI426222305
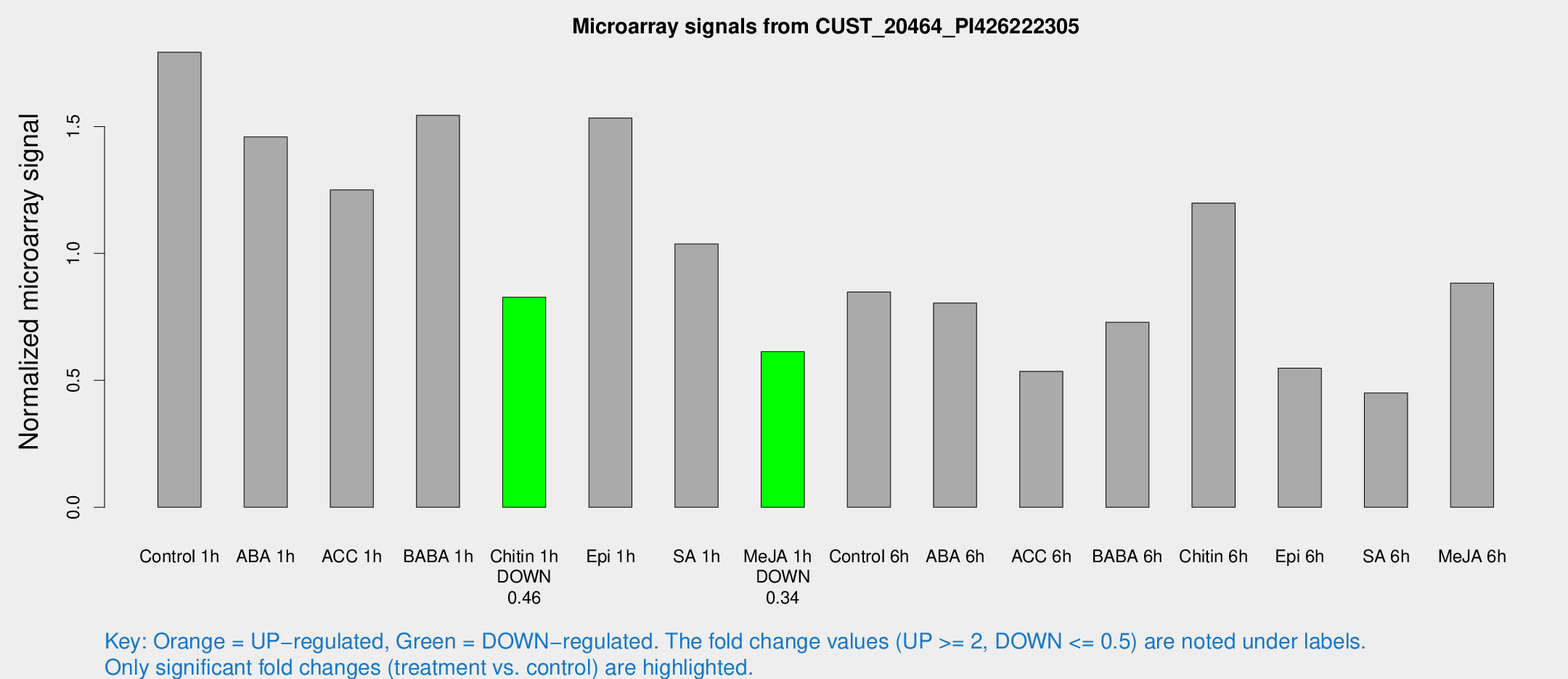
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 68.8287 | 6.92174 | 1.79258 | 0.136121 |
| ABA 1h | 49.6556 | 5.46364 | 1.45892 | 0.215697 |
| ACC 1h | 51.9866 | 11.5982 | 1.25032 | 0.237401 |
| BABA 1h | 58.9356 | 11.3325 | 1.54429 | 0.172313 |
| Chitin 1h | 29.0009 | 4.64348 | 0.827773 | 0.131109 |
| Epi 1h | 51.5794 | 7.82826 | 1.53344 | 0.249208 |
| SA 1h | 40.5974 | 4.03876 | 1.03729 | 0.103352 |
| Me-JA 1h | 20.6849 | 5.33142 | 0.613033 | 0.158064 |
| Control 6h | 36.0277 | 10.3448 | 0.847985 | 0.226838 |
| ABA 6h | 33.0756 | 4.09663 | 0.804887 | 0.102271 |
| ACC 6h | 23.5203 | 4.40034 | 0.535546 | 0.100954 |
| BABA 6h | 31.4506 | 4.21855 | 0.729236 | 0.0995116 |
| Chitin 6h | 49.7769 | 8.20774 | 1.19827 | 0.154307 |
| Epi 6h | 26.6138 | 8.17464 | 0.548486 | 0.187421 |
| SA 6h | 18.5357 | 5.86781 | 0.450202 | 0.118068 |
| Me-JA 6h | 38.5611 | 11.973 | 0.883533 | 0.319148 |
Source Transcript PGSC0003DMT400021305 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G18200.1 | +1 | 2e-65 | 213 | 123/184 (67%) | UTP:galactose-1-phosphate uridylyltransferases;ribose-5-phosphate adenylyltransferases | chr5:6015270-6016555 FORWARD LENGTH=351 |
