Probe CUST_19301_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19301_PI426222305 | JHI_St_60k_v1 | DMT400064585 | AAACTAACGGCATACTCTCAGCAGAAATGGATCTAGAGAAGTATGCAGGACCAGATTCAG |
All Microarray Probes Designed to Gene DMG400025098
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19248_PI426222305 | JHI_St_60k_v1 | DMT400064584 | AACATAGTTGTTGAAAAGGGTCGAGCTGTTGGGGTCAAACTAAGAGGTGGCCAAGATCTA |
| CUST_19280_PI426222305 | JHI_St_60k_v1 | DMT400064583 | TGCATCTGCATTTGGGTTTTGATGCAGAGGATCTAGAGAAGTATGCAGGACCAGATTCAG |
| CUST_19301_PI426222305 | JHI_St_60k_v1 | DMT400064585 | AAACTAACGGCATACTCTCAGCAGAAATGGATCTAGAGAAGTATGCAGGACCAGATTCAG |
| CUST_19402_PI426222305 | JHI_St_60k_v1 | DMT400064586 | AAGAATAATGTGCAGGCAAGCCAGAAGCAGATATCGTTGTTATTGGGAGCGGTATAGGTG |
Microarray Signals from CUST_19301_PI426222305
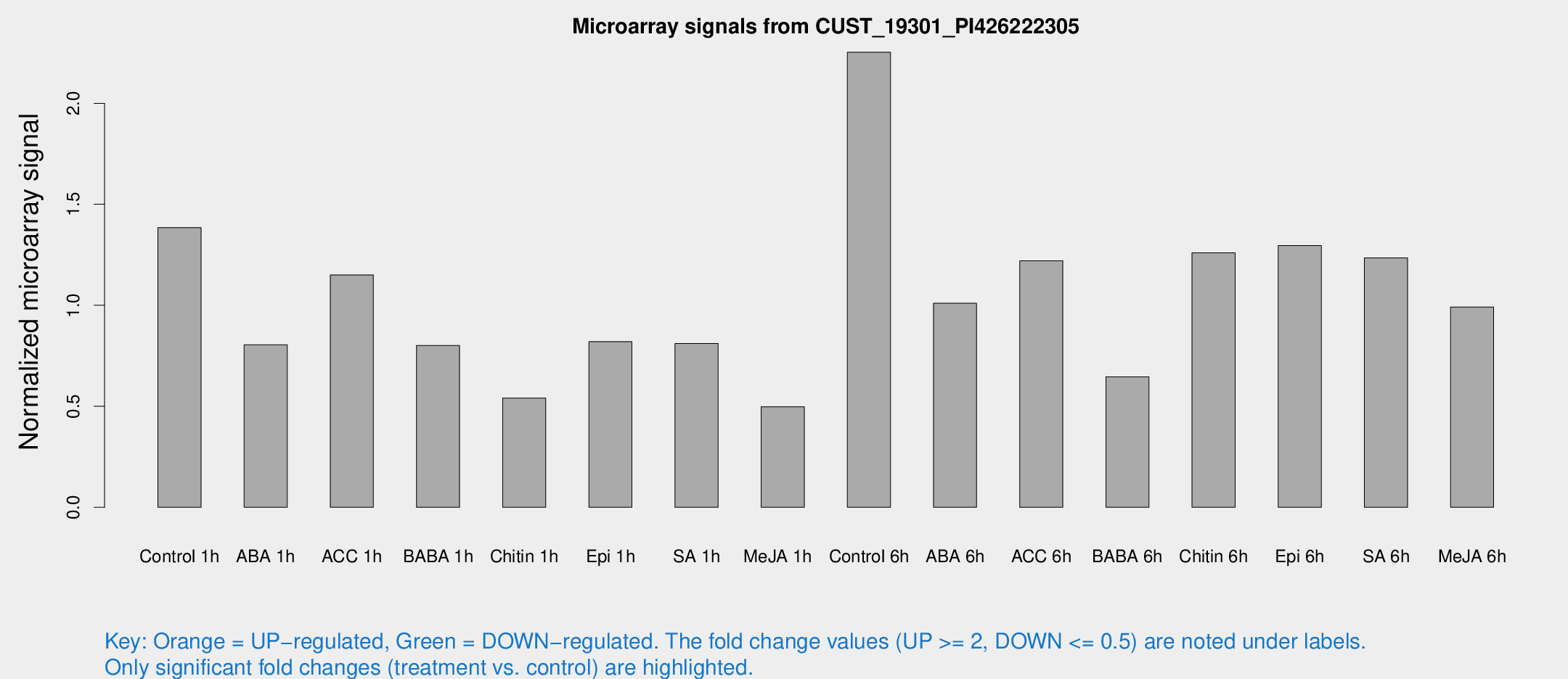
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 37.9819 | 3.92526 | 1.38469 | 0.143722 |
| ABA 1h | 24.8942 | 12.6324 | 0.804654 | 0.511556 |
| ACC 1h | 34.168 | 8.04416 | 1.1501 | 0.219537 |
| BABA 1h | 22.2595 | 5.5633 | 0.800997 | 0.16123 |
| Chitin 1h | 13.4427 | 3.44303 | 0.540811 | 0.142273 |
| Epi 1h | 20.8168 | 5.05029 | 0.82015 | 0.231299 |
| SA 1h | 24.6955 | 7.44917 | 0.810979 | 0.207182 |
| Me-JA 1h | 12.8089 | 4.54196 | 0.497703 | 0.181212 |
| Control 6h | 65.2405 | 14.2287 | 2.25277 | 0.373646 |
| ABA 6h | 31.5894 | 9.20793 | 1.01016 | 0.293174 |
| ACC 6h | 40.9073 | 9.7389 | 1.22044 | 0.247523 |
| BABA 6h | 22.3746 | 7.36765 | 0.645431 | 0.2865 |
| Chitin 6h | 38.8251 | 10.0747 | 1.25967 | 0.277279 |
| Epi 6h | 45.0025 | 16.6231 | 1.29549 | 0.474261 |
| SA 6h | 35.6267 | 8.2607 | 1.23485 | 0.195931 |
| Me-JA 6h | 27.5774 | 4.12114 | 0.991732 | 0.147566 |
Source Transcript PGSC0003DMT400064585 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G57770.1 | +1 | 0.0 | 879 | 452/539 (84%) | FAD/NAD(P)-binding oxidoreductase family protein | chr1:21395254-21398135 FORWARD LENGTH=574 |
