Probe CUST_19280_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19280_PI426222305 | JHI_St_60k_v1 | DMT400064583 | TGCATCTGCATTTGGGTTTTGATGCAGAGGATCTAGAGAAGTATGCAGGACCAGATTCAG |
All Microarray Probes Designed to Gene DMG400025098
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19248_PI426222305 | JHI_St_60k_v1 | DMT400064584 | AACATAGTTGTTGAAAAGGGTCGAGCTGTTGGGGTCAAACTAAGAGGTGGCCAAGATCTA |
| CUST_19280_PI426222305 | JHI_St_60k_v1 | DMT400064583 | TGCATCTGCATTTGGGTTTTGATGCAGAGGATCTAGAGAAGTATGCAGGACCAGATTCAG |
| CUST_19301_PI426222305 | JHI_St_60k_v1 | DMT400064585 | AAACTAACGGCATACTCTCAGCAGAAATGGATCTAGAGAAGTATGCAGGACCAGATTCAG |
| CUST_19402_PI426222305 | JHI_St_60k_v1 | DMT400064586 | AAGAATAATGTGCAGGCAAGCCAGAAGCAGATATCGTTGTTATTGGGAGCGGTATAGGTG |
Microarray Signals from CUST_19280_PI426222305
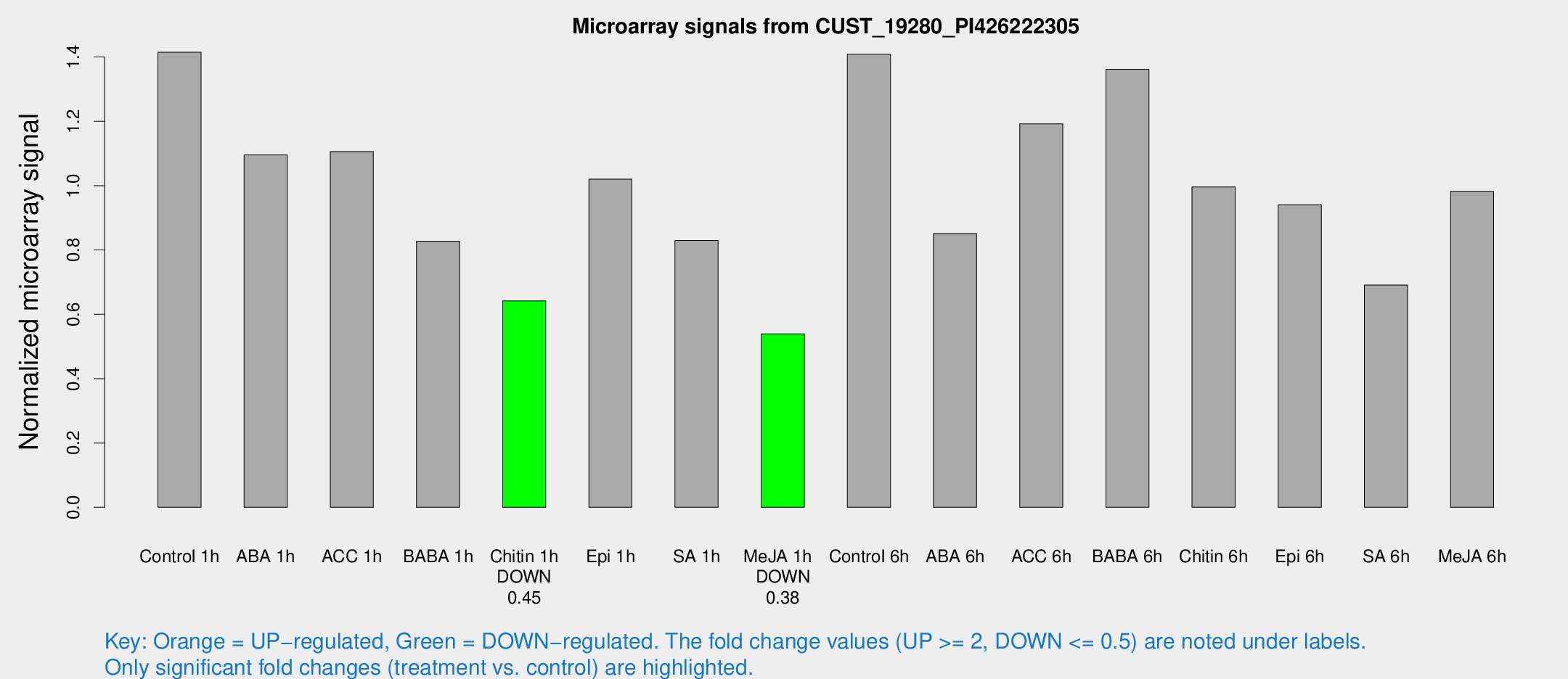
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 99.6852 | 11.0892 | 1.41469 | 0.0973698 |
| ABA 1h | 69.4988 | 11.166 | 1.09565 | 0.279811 |
| ACC 1h | 83.1751 | 16.9632 | 1.106 | 0.176201 |
| BABA 1h | 60.1901 | 14.9744 | 0.827314 | 0.167074 |
| Chitin 1h | 40.2308 | 4.1672 | 0.641486 | 0.0808955 |
| Epi 1h | 62.655 | 8.91317 | 1.01978 | 0.164393 |
| SA 1h | 60.3376 | 8.03509 | 0.829556 | 0.102026 |
| Me-JA 1h | 31.469 | 5.1023 | 0.539354 | 0.0712492 |
| Control 6h | 112.104 | 36.1099 | 1.40869 | 0.452717 |
| ABA 6h | 67.3376 | 18.0253 | 0.850997 | 0.22144 |
| ACC 6h | 96.5092 | 10.8603 | 1.19215 | 0.119085 |
| BABA 6h | 149.624 | 81.3788 | 1.36149 | 1.25041 |
| Chitin 6h | 76.3411 | 14.6446 | 0.996065 | 0.156557 |
| Epi 6h | 75.8254 | 12.6995 | 0.940666 | 0.185818 |
| SA 6h | 57.5163 | 20.5267 | 0.69069 | 0.275313 |
| Me-JA 6h | 70.5223 | 12.463 | 0.982273 | 0.134554 |
Source Transcript PGSC0003DMT400064583 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G57770.1 | +2 | 8e-132 | 406 | 207/237 (87%) | FAD/NAD(P)-binding oxidoreductase family protein | chr1:21395254-21398135 FORWARD LENGTH=574 |
