Probe CUST_19116_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19116_PI426222305 | JHI_St_60k_v1 | DMT400044692 | GGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTTGAA |
All Microarray Probes Designed to Gene DMG400017338
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19028_PI426222305 | JHI_St_60k_v1 | DMT400044691 | TGCTGTTTTAAAATGGTTTATAGGCCGTGCTCAGGATGCTCAGCGAGATTGCAAGTATTG |
| CUST_18952_PI426222305 | JHI_St_60k_v1 | DMT400044693 | GGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTTGAA |
| CUST_18950_PI426222305 | JHI_St_60k_v1 | DMT400044690 | CTCGGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTT |
| CUST_19116_PI426222305 | JHI_St_60k_v1 | DMT400044692 | GGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTTGAA |
Microarray Signals from CUST_19116_PI426222305
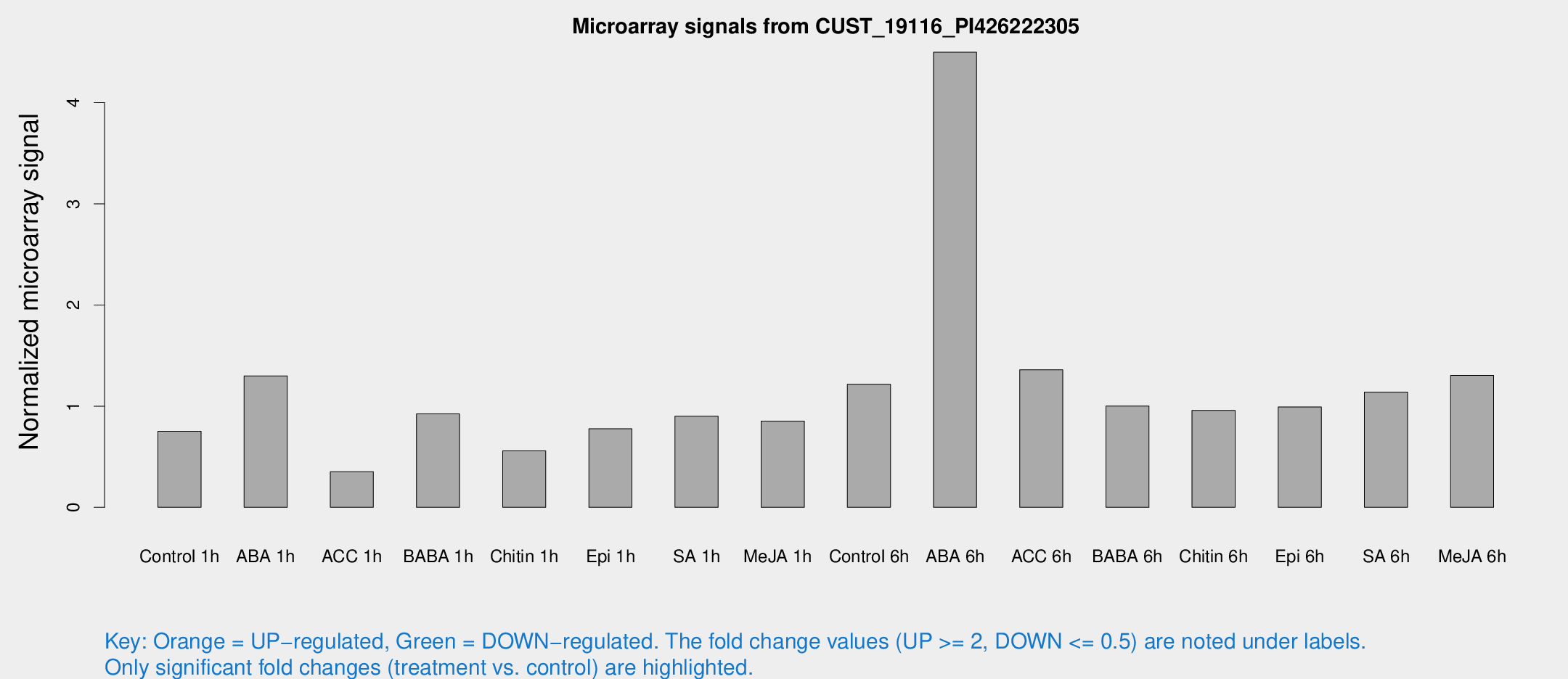
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 57.9093 | 4.8593 | 0.752026 | 0.0640298 |
| ABA 1h | 88.6804 | 8.35196 | 1.29808 | 0.0904265 |
| ACC 1h | 33.7721 | 15.6594 | 0.351724 | 0.157309 |
| BABA 1h | 72.2178 | 16.8771 | 0.923407 | 0.145854 |
| Chitin 1h | 41.9916 | 11.3521 | 0.559029 | 0.176823 |
| Epi 1h | 54.5533 | 11.8594 | 0.776376 | 0.194283 |
| SA 1h | 71.7915 | 8.68901 | 0.900533 | 0.0705871 |
| Me-JA 1h | 55.2458 | 11.1163 | 0.851623 | 0.0954246 |
| Control 6h | 97.2197 | 19.5086 | 1.21666 | 0.300417 |
| ABA 6h | 391.757 | 91.8297 | 4.49893 | 1.0455 |
| ACC 6h | 119.86 | 8.3775 | 1.35902 | 0.219022 |
| BABA 6h | 86.3709 | 6.99459 | 1.0015 | 0.0765177 |
| Chitin 6h | 80.2846 | 13.3934 | 0.958543 | 0.146251 |
| Epi 6h | 87.144 | 12.1504 | 0.991742 | 0.0850751 |
| SA 6h | 103.237 | 34.8587 | 1.13952 | 0.436913 |
| Me-JA 6h | 102.727 | 19.6364 | 1.30348 | 0.162388 |
Source Transcript PGSC0003DMT400044692 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G58750.1 | +3 | 0.0 | 818 | 410/504 (81%) | citrate synthase 2 | chr3:21724564-21727458 REVERSE LENGTH=514 |
