Probe CUST_18950_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18950_PI426222305 | JHI_St_60k_v1 | DMT400044690 | CTCGGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTT |
All Microarray Probes Designed to Gene DMG400017338
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19028_PI426222305 | JHI_St_60k_v1 | DMT400044691 | TGCTGTTTTAAAATGGTTTATAGGCCGTGCTCAGGATGCTCAGCGAGATTGCAAGTATTG |
| CUST_18952_PI426222305 | JHI_St_60k_v1 | DMT400044693 | GGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTTGAA |
| CUST_18950_PI426222305 | JHI_St_60k_v1 | DMT400044690 | CTCGGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTT |
| CUST_19116_PI426222305 | JHI_St_60k_v1 | DMT400044692 | GGATGGGCATAGTTTCGGCAATTCAAATCAAAATTCGAACTTTCTGATATTTGGTTTGAA |
Microarray Signals from CUST_18950_PI426222305
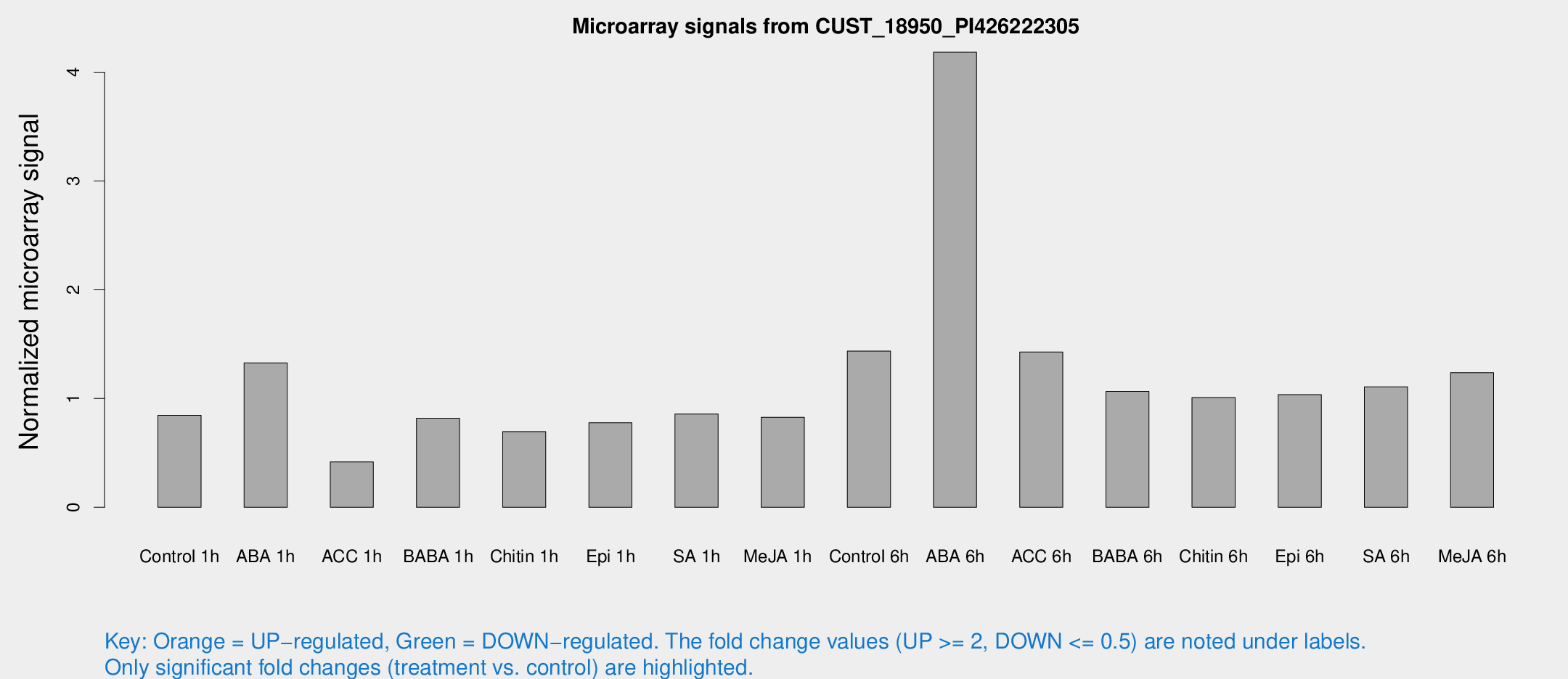
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 94.8175 | 10.1633 | 0.846144 | 0.124424 |
| ABA 1h | 138.631 | 33.2634 | 1.32795 | 0.311296 |
| ACC 1h | 57.8114 | 20.9933 | 0.418107 | 0.194623 |
| BABA 1h | 89.262 | 12.8593 | 0.81881 | 0.0698778 |
| Chitin 1h | 74.4604 | 17.6271 | 0.695905 | 0.211496 |
| Epi 1h | 77.4532 | 14.4613 | 0.777101 | 0.154228 |
| SA 1h | 100.74 | 16.9451 | 0.856711 | 0.111931 |
| Me-JA 1h | 76.6766 | 12.2712 | 0.8277 | 0.0764235 |
| Control 6h | 166.78 | 36.7278 | 1.43519 | 0.324544 |
| ABA 6h | 521.211 | 111.603 | 4.18324 | 0.859831 |
| ACC 6h | 182.201 | 11.9799 | 1.42727 | 0.269966 |
| BABA 6h | 132.367 | 8.54751 | 1.06553 | 0.0687223 |
| Chitin 6h | 119.123 | 7.8736 | 1.00834 | 0.0665532 |
| Epi 6h | 130.648 | 12.5124 | 1.03536 | 0.0674911 |
| SA 6h | 139.479 | 44.5113 | 1.10761 | 0.333169 |
| Me-JA 6h | 139.696 | 21.7353 | 1.23691 | 0.0924791 |
Source Transcript PGSC0003DMT400044690 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G58750.1 | +1 | 3e-90 | 279 | 135/165 (82%) | citrate synthase 2 | chr3:21724564-21727458 REVERSE LENGTH=514 |
