Probe CUST_17900_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17900_PI426222305 | JHI_St_60k_v1 | DMT400006303 | CGTTTCTATTGGAGAACCGATTTGACGTATAATTGCAGGAATAACACCATTTGTGACTCT |
All Microarray Probes Designed to Gene DMG400002462
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17900_PI426222305 | JHI_St_60k_v1 | DMT400006303 | CGTTTCTATTGGAGAACCGATTTGACGTATAATTGCAGGAATAACACCATTTGTGACTCT |
| CUST_17859_PI426222305 | JHI_St_60k_v1 | DMT400006302 | GGAGCAGAGTCCTCCCTAGTACTTTTCGTTATGCTTTTGTTTGTATTTTGTTCCTTTGAT |
| CUST_17937_PI426222305 | JHI_St_60k_v1 | DMT400006305 | GTTGAGTAATTGGGATGTTTTTAGTCATACTAAAGCCCAAATGAAGAGTAGCAGTACTAA |
Microarray Signals from CUST_17900_PI426222305
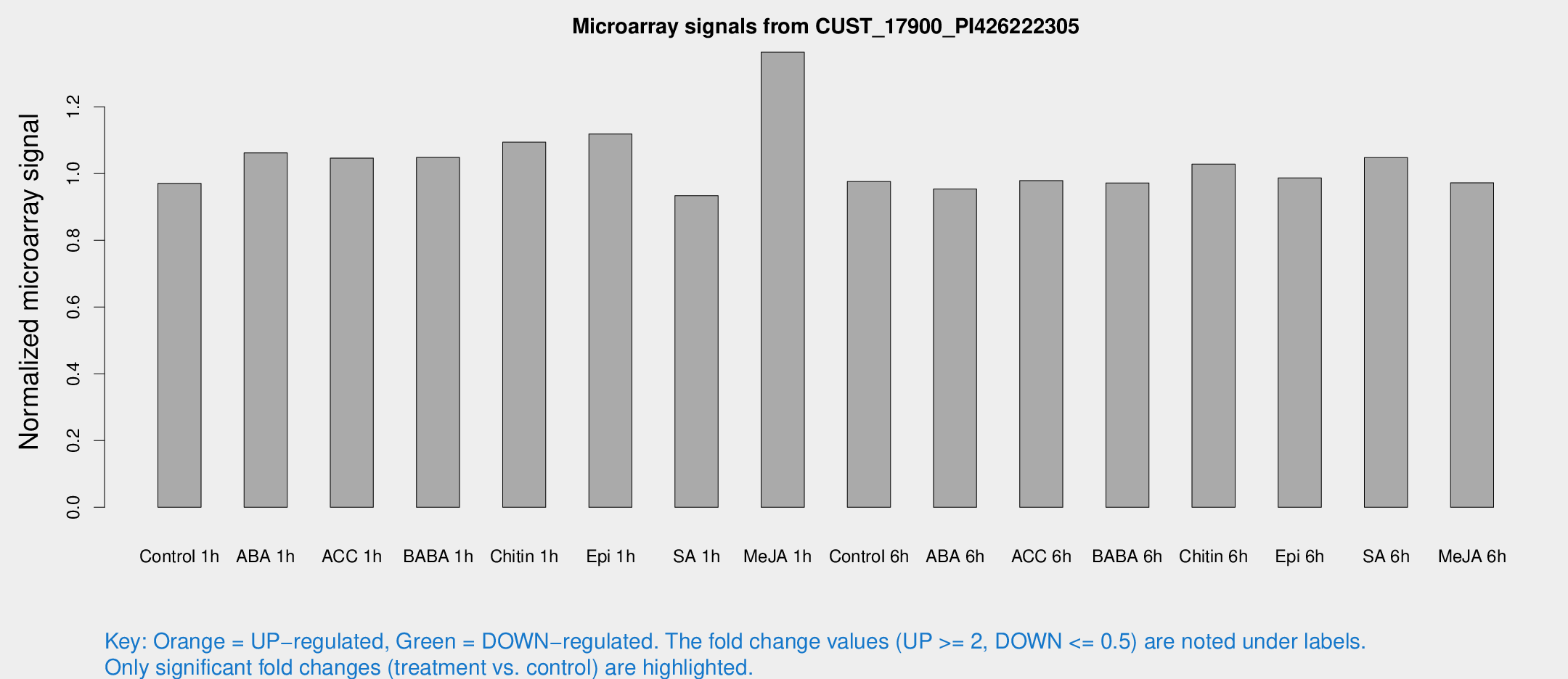
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.10744 | 2.96036 | 0.970626 | 0.554124 |
| ABA 1h | 4.87093 | 2.82527 | 1.0619 | 0.598602 |
| ACC 1h | 5.76593 | 3.36255 | 1.04655 | 0.60667 |
| BABA 1h | 5.39715 | 3.12984 | 1.04839 | 0.607079 |
| Chitin 1h | 5.06135 | 2.94616 | 1.09415 | 0.609051 |
| Epi 1h | 5.03071 | 2.91896 | 1.11879 | 0.630432 |
| SA 1h | 5.11566 | 2.96276 | 0.93366 | 0.540668 |
| Me-JA 1h | 5.9305 | 3.01192 | 1.3638 | 0.697126 |
| Control 6h | 5.21239 | 3.0228 | 0.976167 | 0.564995 |
| ABA 6h | 5.41471 | 3.14199 | 0.953989 | 0.552936 |
| ACC 6h | 6.12775 | 3.63382 | 0.979229 | 0.567043 |
| BABA 6h | 5.81353 | 3.37312 | 0.972183 | 0.563118 |
| Chitin 6h | 5.84305 | 3.35081 | 1.02823 | 0.589737 |
| Epi 6h | 5.98924 | 3.50099 | 0.986912 | 0.571477 |
| SA 6h | 5.53045 | 3.20375 | 1.04811 | 0.607005 |
| Me-JA 6h | 5.16366 | 2.99407 | 0.972435 | 0.563417 |
Source Transcript PGSC0003DMT400006303 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G60140.1 | +1 | 9e-67 | 159 | 87/222 (39%) | Glycosyl hydrolase superfamily protein | chr3:22216753-22220710 FORWARD LENGTH=577 |
