Probe CUST_17859_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17859_PI426222305 | JHI_St_60k_v1 | DMT400006302 | GGAGCAGAGTCCTCCCTAGTACTTTTCGTTATGCTTTTGTTTGTATTTTGTTCCTTTGAT |
All Microarray Probes Designed to Gene DMG400002462
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17900_PI426222305 | JHI_St_60k_v1 | DMT400006303 | CGTTTCTATTGGAGAACCGATTTGACGTATAATTGCAGGAATAACACCATTTGTGACTCT |
| CUST_17859_PI426222305 | JHI_St_60k_v1 | DMT400006302 | GGAGCAGAGTCCTCCCTAGTACTTTTCGTTATGCTTTTGTTTGTATTTTGTTCCTTTGAT |
| CUST_17937_PI426222305 | JHI_St_60k_v1 | DMT400006305 | GTTGAGTAATTGGGATGTTTTTAGTCATACTAAAGCCCAAATGAAGAGTAGCAGTACTAA |
Microarray Signals from CUST_17859_PI426222305
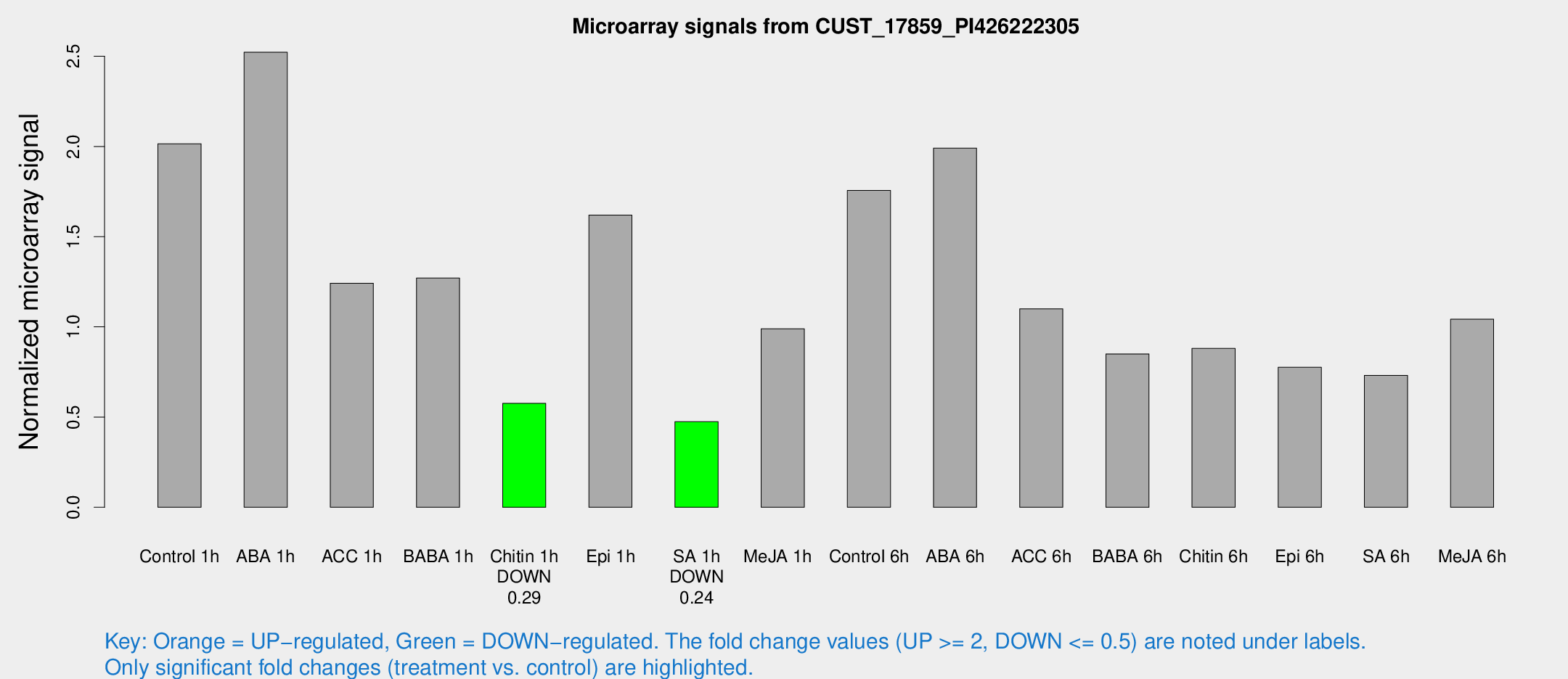
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 22.9114 | 4.8724 | 2.01515 | 0.326848 |
| ABA 1h | 28.1229 | 11.4164 | 2.52227 | 1.13419 |
| ACC 1h | 16.6574 | 6.58328 | 1.24213 | 0.538432 |
| BABA 1h | 15.2945 | 5.92786 | 1.27049 | 0.413781 |
| Chitin 1h | 5.53762 | 3.0929 | 0.576236 | 0.315631 |
| Epi 1h | 15.7221 | 3.21171 | 1.61985 | 0.36665 |
| SA 1h | 5.28201 | 3.06417 | 0.474596 | 0.275287 |
| Me-JA 1h | 11.7964 | 6.62042 | 0.98961 | 0.523253 |
| Control 6h | 25.8488 | 11.097 | 1.75609 | 1.17736 |
| ABA 6h | 23.5195 | 3.73775 | 1.99102 | 0.317634 |
| ACC 6h | 13.9174 | 3.9718 | 1.10024 | 0.313658 |
| BABA 6h | 13.5621 | 7.31412 | 0.850114 | 0.510649 |
| Chitin 6h | 11.8672 | 4.81774 | 0.880548 | 0.409586 |
| Epi 6h | 10.2822 | 3.71592 | 0.775968 | 0.33207 |
| SA 6h | 8.35029 | 3.38652 | 0.731561 | 0.336904 |
| Me-JA 6h | 15.7717 | 9.1923 | 1.04257 | 0.620862 |
Source Transcript PGSC0003DMT400006302 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G61820.1 | +2 | 3e-128 | 229 | 113/263 (43%) | beta glucosidase 46 | chr1:22835452-22838444 FORWARD LENGTH=516 |
