Probe CUST_1613_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1613_PI426222305 | JHI_St_60k_v1 | DMT400043632 | GCAGCTTTTGACAATCATTTCTTCTGGAAATCCAAAGGAAGAAACATTTCCTAGATCCAG |
All Microarray Probes Designed to Gene DMG400016941
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1579_PI426222305 | JHI_St_60k_v1 | DMT400043631 | GCAGCTTTTGACAATCATTTCTTCTGGAAATCCAAAGGAAGAAACATTTCCTAGATCCAG |
| CUST_1613_PI426222305 | JHI_St_60k_v1 | DMT400043632 | GCAGCTTTTGACAATCATTTCTTCTGGAAATCCAAAGGAAGAAACATTTCCTAGATCCAG |
Microarray Signals from CUST_1613_PI426222305
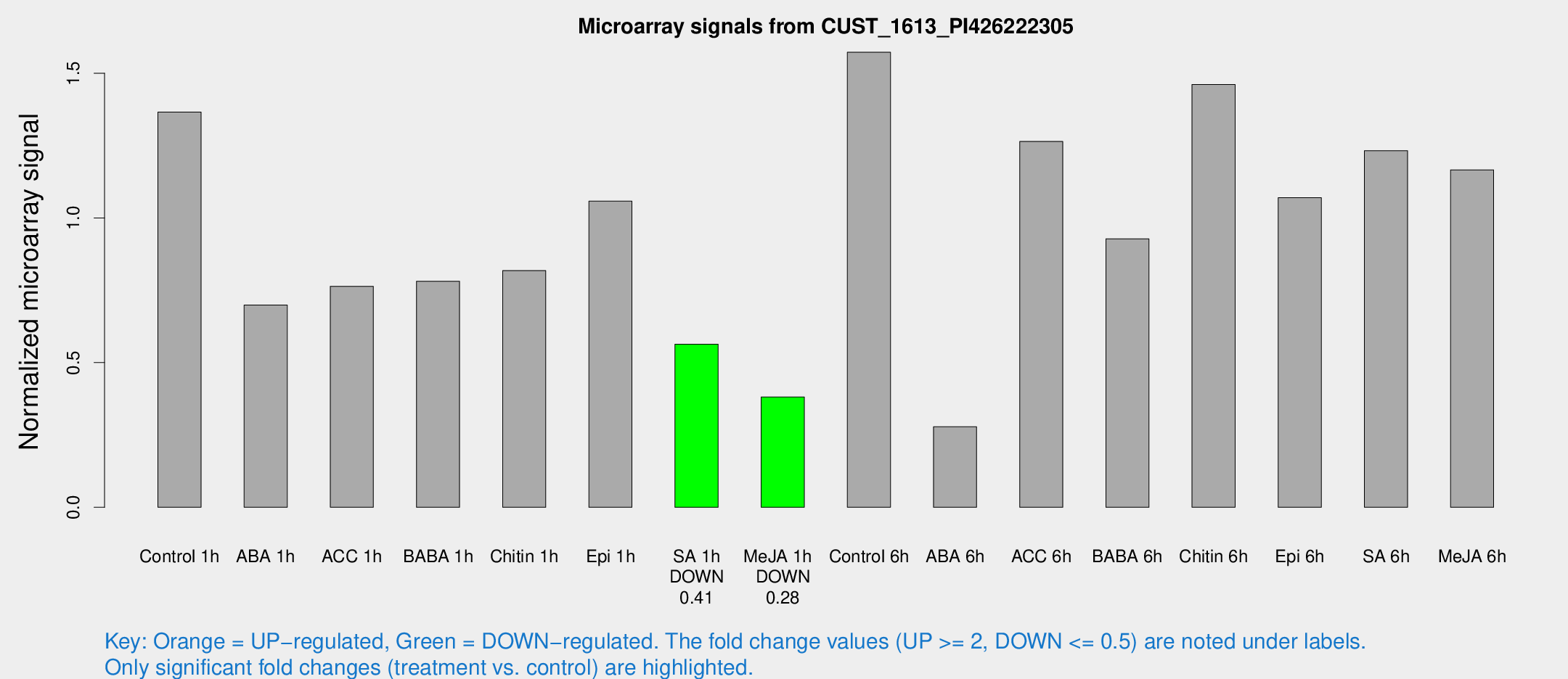
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 94.8469 | 9.17652 | 1.36555 | 0.0911847 |
| ABA 1h | 46.7149 | 14.6134 | 0.699055 | 0.254311 |
| ACC 1h | 70.1381 | 26.7292 | 0.763723 | 0.472311 |
| BABA 1h | 56.8214 | 15.0526 | 0.781163 | 0.176646 |
| Chitin 1h | 50.7375 | 4.27735 | 0.817943 | 0.0687437 |
| Epi 1h | 63.53 | 5.99062 | 1.05774 | 0.102895 |
| SA 1h | 40.5575 | 5.38722 | 0.56372 | 0.0646125 |
| Me-JA 1h | 22.189 | 4.18863 | 0.381343 | 0.0617576 |
| Control 6h | 122.425 | 43.4885 | 1.57244 | 0.486817 |
| ABA 6h | 21.4972 | 5.1203 | 0.278363 | 0.0638449 |
| ACC 6h | 107.999 | 26.9188 | 1.26422 | 0.341141 |
| BABA 6h | 77.7414 | 19.8327 | 0.927726 | 0.267337 |
| Chitin 6h | 110.037 | 18.2257 | 1.46112 | 0.228563 |
| Epi 6h | 86.6942 | 18.7073 | 1.0697 | 0.256701 |
| SA 6h | 95.1091 | 35.1422 | 1.23207 | 0.443397 |
| Me-JA 6h | 90.0376 | 29.4389 | 1.16605 | 0.333383 |
Source Transcript PGSC0003DMT400043632 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc06g051460.2 | +3 | 0.0 | 1682 | 937/1065 (88%) | genomic_reference:SL2.50ch06 gene_region:31369126-31374317 transcript_region:SL2.50ch06:31369126..31374317- functional_description:ATP-dependent chaperone ClpB (AHRD V1 *--- C0GC24_9FIRM) |
| TAIR PP10 | AT2G29970.1 | +3 | 0.0 | 598 | 434/1085 (40%) | Double Clp-N motif-containing P-loop nucleoside triphosphate hydrolases superfamily protein | chr2:12776601-12779784 FORWARD LENGTH=1002 |
