Probe CUST_15498_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15498_PI426222305 | JHI_St_60k_v1 | DMT400037471 | TAGTTTGCTTATTAATATCTCTATTGACAGACTGTATACGATCACGACACATTCAGCATG |
All Microarray Probes Designed to Gene DMG400014458
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15498_PI426222305 | JHI_St_60k_v1 | DMT400037471 | TAGTTTGCTTATTAATATCTCTATTGACAGACTGTATACGATCACGACACATTCAGCATG |
| CUST_15447_PI426222305 | JHI_St_60k_v1 | DMT400037470 | GCACAAGGTTTGTAGCTTAGTGGATATGGGGTTTGAAAGTTTATCTTTTTAGTCATGTTA |
| CUST_15497_PI426222305 | JHI_St_60k_v1 | DMT400037472 | GCACAAGGTTTGTAGCTTAGTGGATATGGGGTTTGAAAGTTTATCTTTTTAGTCATGTTA |
Microarray Signals from CUST_15498_PI426222305
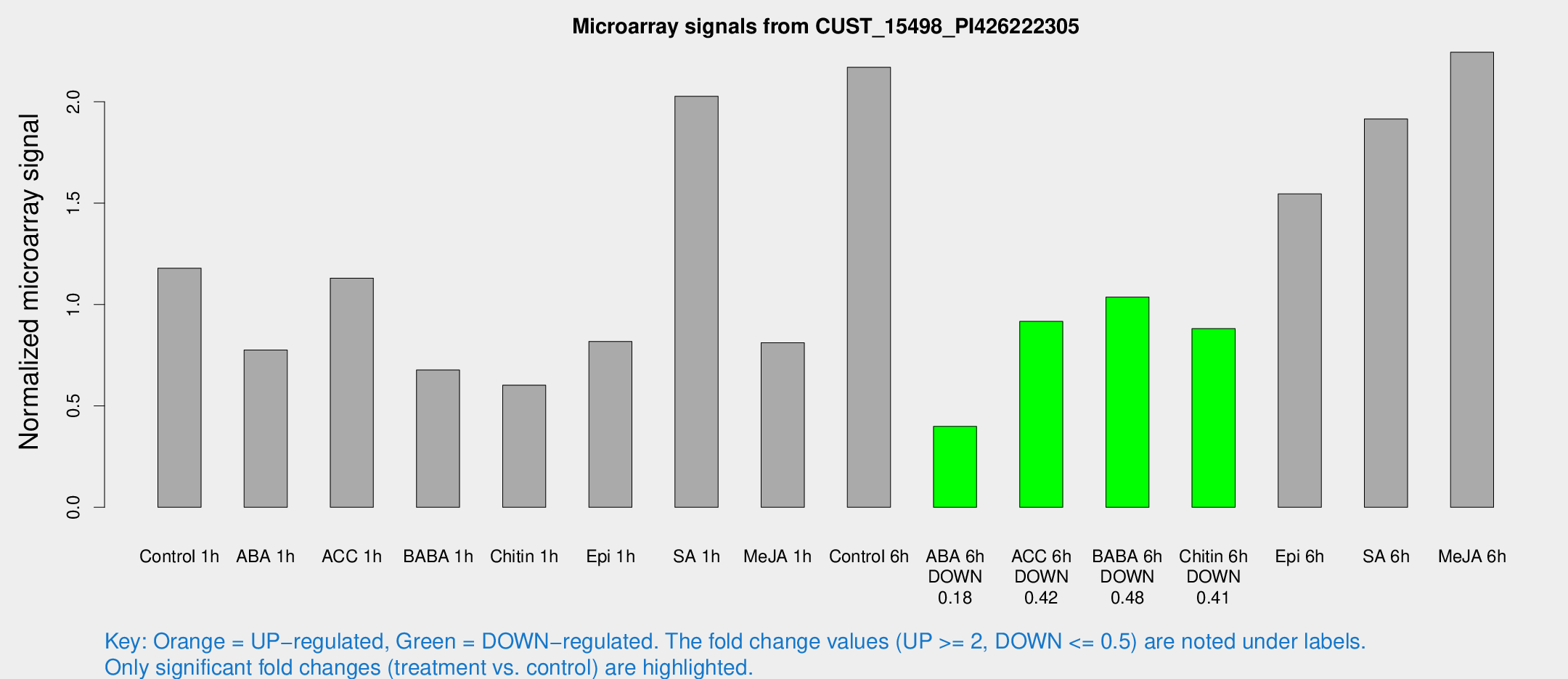
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 143.547 | 51.8101 | 1.17964 | 0.340766 |
| ABA 1h | 81.5003 | 25.079 | 0.775897 | 0.237172 |
| ACC 1h | 131.673 | 29.9527 | 1.13033 | 0.22058 |
| BABA 1h | 83.4771 | 28.4203 | 0.677085 | 0.252949 |
| Chitin 1h | 61.4464 | 13.9072 | 0.602307 | 0.0854809 |
| Epi 1h | 78.6483 | 12.2092 | 0.818012 | 0.119742 |
| SA 1h | 238.049 | 57.3196 | 2.02725 | 0.353606 |
| Me-JA 1h | 77.7046 | 23.1326 | 0.811987 | 0.167494 |
| Control 6h | 249.878 | 54.8632 | 2.17036 | 0.354827 |
| ABA 6h | 46.4157 | 4.92611 | 0.398855 | 0.0353247 |
| ACC 6h | 117.041 | 19.5414 | 0.91697 | 0.194076 |
| BABA 6h | 126.215 | 8.57019 | 1.03726 | 0.0659138 |
| Chitin 6h | 108.751 | 28.3858 | 0.880942 | 0.22843 |
| Epi 6h | 193.785 | 30.0508 | 1.54565 | 0.151285 |
| SA 6h | 276.279 | 111.131 | 1.91569 | 1.19383 |
| Me-JA 6h | 247.839 | 38.2698 | 2.24453 | 0.340476 |
Source Transcript PGSC0003DMT400037471 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G17980.1 | +2 | 3e-44 | 156 | 72/94 (77%) | Calcium-dependent lipid-binding (CaLB domain) family protein | chr3:6152417-6153115 FORWARD LENGTH=177 |
