Probe CUST_15447_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15447_PI426222305 | JHI_St_60k_v1 | DMT400037470 | GCACAAGGTTTGTAGCTTAGTGGATATGGGGTTTGAAAGTTTATCTTTTTAGTCATGTTA |
All Microarray Probes Designed to Gene DMG400014458
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15498_PI426222305 | JHI_St_60k_v1 | DMT400037471 | TAGTTTGCTTATTAATATCTCTATTGACAGACTGTATACGATCACGACACATTCAGCATG |
| CUST_15447_PI426222305 | JHI_St_60k_v1 | DMT400037470 | GCACAAGGTTTGTAGCTTAGTGGATATGGGGTTTGAAAGTTTATCTTTTTAGTCATGTTA |
| CUST_15497_PI426222305 | JHI_St_60k_v1 | DMT400037472 | GCACAAGGTTTGTAGCTTAGTGGATATGGGGTTTGAAAGTTTATCTTTTTAGTCATGTTA |
Microarray Signals from CUST_15447_PI426222305
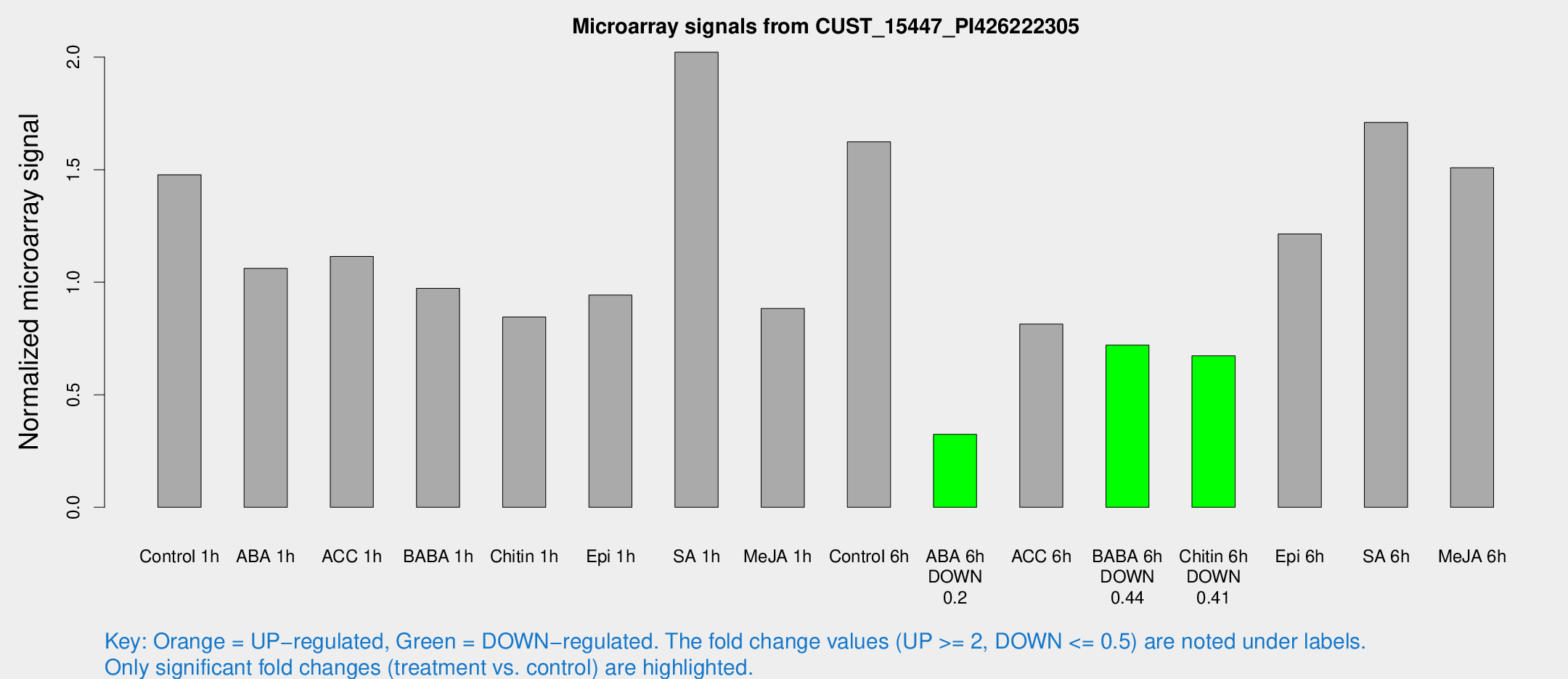
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5596.99 | 638.64 | 1.47748 | 0.0853064 |
| ABA 1h | 3630.28 | 672.474 | 1.06132 | 0.186536 |
| ACC 1h | 4521.76 | 1063.47 | 1.1144 | 0.20758 |
| BABA 1h | 3789.86 | 937.676 | 0.97282 | 0.188705 |
| Chitin 1h | 2847.71 | 164.699 | 0.845405 | 0.0558098 |
| Epi 1h | 3146.56 | 511.983 | 0.942596 | 0.160239 |
| SA 1h | 7902.81 | 1007.57 | 2.02189 | 0.144608 |
| Me-JA 1h | 2752.82 | 389.136 | 0.883755 | 0.0510341 |
| Control 6h | 6342.14 | 1261.38 | 1.62404 | 0.229004 |
| ABA 6h | 1296.51 | 103.448 | 0.32386 | 0.0187167 |
| ACC 6h | 3524.08 | 336.83 | 0.813906 | 0.132584 |
| BABA 6h | 3021.61 | 174.809 | 0.720361 | 0.0501405 |
| Chitin 6h | 2702.61 | 243.846 | 0.673327 | 0.0513792 |
| Epi 6h | 5266.37 | 857.945 | 1.21428 | 0.113214 |
| SA 6h | 7638.37 | 2616.34 | 1.71003 | 0.685727 |
| Me-JA 6h | 5754.84 | 915.055 | 1.50895 | 0.170614 |
Source Transcript PGSC0003DMT400037470 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G17980.1 | +2 | 2e-78 | 242 | 130/168 (77%) | Calcium-dependent lipid-binding (CaLB domain) family protein | chr3:6152417-6153115 FORWARD LENGTH=177 |
