Probe CUST_14735_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14735_PI426222305 | JHI_St_60k_v1 | DMT400066125 | CACAACATCTGCTCACTTCATTGTGGTCATCGCTTCTTTTGCAGGTATGGTTGTTACATC |
All Microarray Probes Designed to Gene DMG400025737
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14678_PI426222305 | JHI_St_60k_v1 | DMT400066123 | TGTGGGAGTTCCTGTTGATCCCCTTGCTTTTGCTGGACTTTTAGGTATGGTTGTTACATC |
| CUST_14735_PI426222305 | JHI_St_60k_v1 | DMT400066125 | CACAACATCTGCTCACTTCATTGTGGTCATCGCTTCTTTTGCAGGTATGGTTGTTACATC |
| CUST_14616_PI426222305 | JHI_St_60k_v1 | DMT400066122 | GAACTTAAATGGAGCTGCTTATTCAGTGAGGACTAATTGTGCTTGTATACATGGTTAGGC |
Microarray Signals from CUST_14735_PI426222305
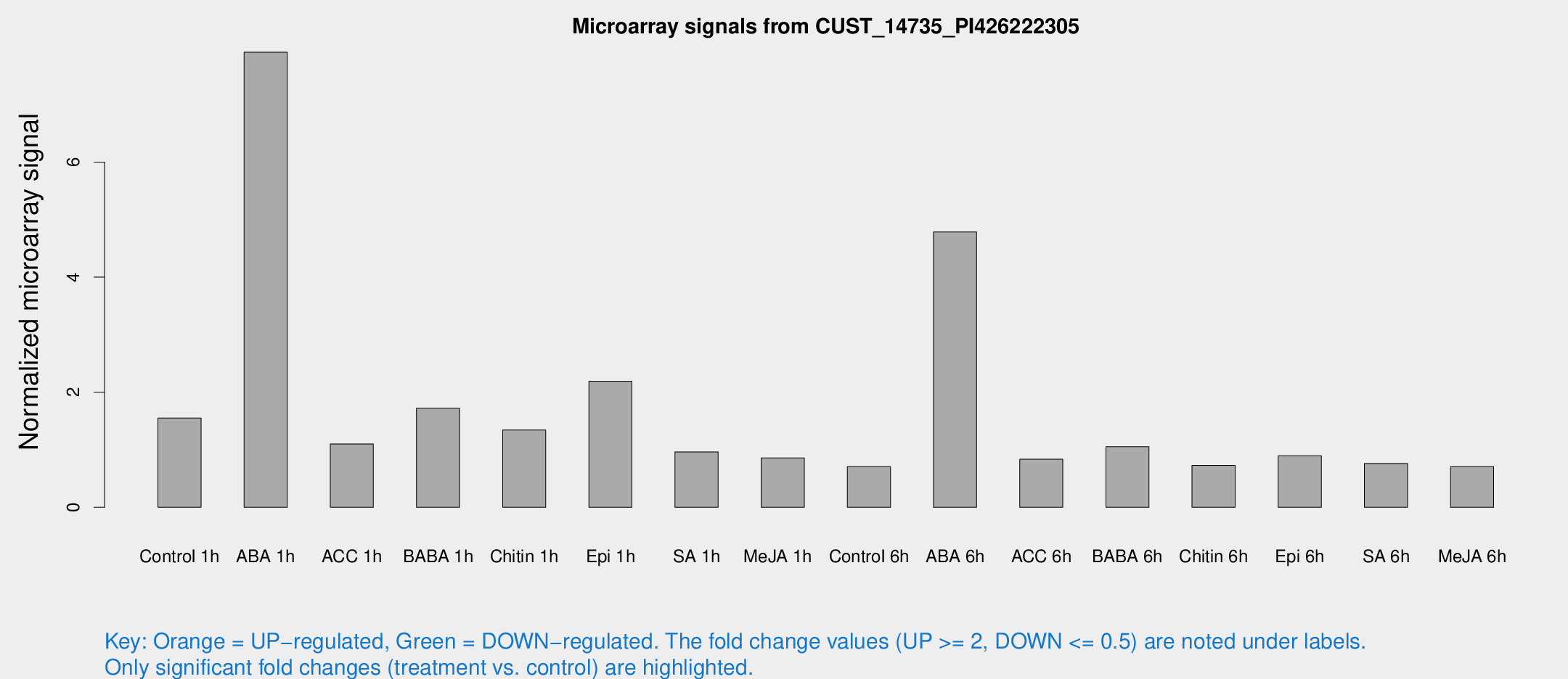
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 16.5048 | 5.10822 | 1.55253 | 0.854215 |
| ABA 1h | 70.0431 | 20.7839 | 7.90844 | 1.72342 |
| ACC 1h | 11.4081 | 4.56671 | 1.10056 | 0.518095 |
| BABA 1h | 18.4968 | 7.05824 | 1.72066 | 0.688452 |
| Chitin 1h | 11.4568 | 3.69729 | 1.34341 | 0.449036 |
| Epi 1h | 17.5599 | 3.70545 | 2.19155 | 0.46339 |
| SA 1h | 9.48526 | 3.66728 | 0.961296 | 0.404235 |
| Me-JA 1h | 6.48678 | 3.76952 | 0.85745 | 0.497225 |
| Control 6h | 6.54243 | 3.79016 | 0.705662 | 0.408646 |
| ABA 6h | 51.9391 | 14.2832 | 4.78504 | 1.51733 |
| ACC 6h | 9.027 | 4.60995 | 0.834782 | 0.424029 |
| BABA 6h | 11.9701 | 4.2193 | 1.05267 | 0.435971 |
| Chitin 6h | 7.1685 | 4.15567 | 0.728001 | 0.421902 |
| Epi 6h | 9.65481 | 4.66644 | 0.895895 | 0.451025 |
| SA 6h | 6.9711 | 4.04432 | 0.761627 | 0.441619 |
| Me-JA 6h | 6.50749 | 3.7701 | 0.706351 | 0.409019 |
Source Transcript PGSC0003DMT400066125 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G17870.1 | +1 | 3e-41 | 158 | 101/123 (82%) | ethylene-dependent gravitropism-deficient and yellow-green-like 3 | chr1:6150036-6152185 REVERSE LENGTH=573 |
