Probe CUST_14616_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14616_PI426222305 | JHI_St_60k_v1 | DMT400066122 | GAACTTAAATGGAGCTGCTTATTCAGTGAGGACTAATTGTGCTTGTATACATGGTTAGGC |
All Microarray Probes Designed to Gene DMG400025737
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14678_PI426222305 | JHI_St_60k_v1 | DMT400066123 | TGTGGGAGTTCCTGTTGATCCCCTTGCTTTTGCTGGACTTTTAGGTATGGTTGTTACATC |
| CUST_14735_PI426222305 | JHI_St_60k_v1 | DMT400066125 | CACAACATCTGCTCACTTCATTGTGGTCATCGCTTCTTTTGCAGGTATGGTTGTTACATC |
| CUST_14616_PI426222305 | JHI_St_60k_v1 | DMT400066122 | GAACTTAAATGGAGCTGCTTATTCAGTGAGGACTAATTGTGCTTGTATACATGGTTAGGC |
Microarray Signals from CUST_14616_PI426222305
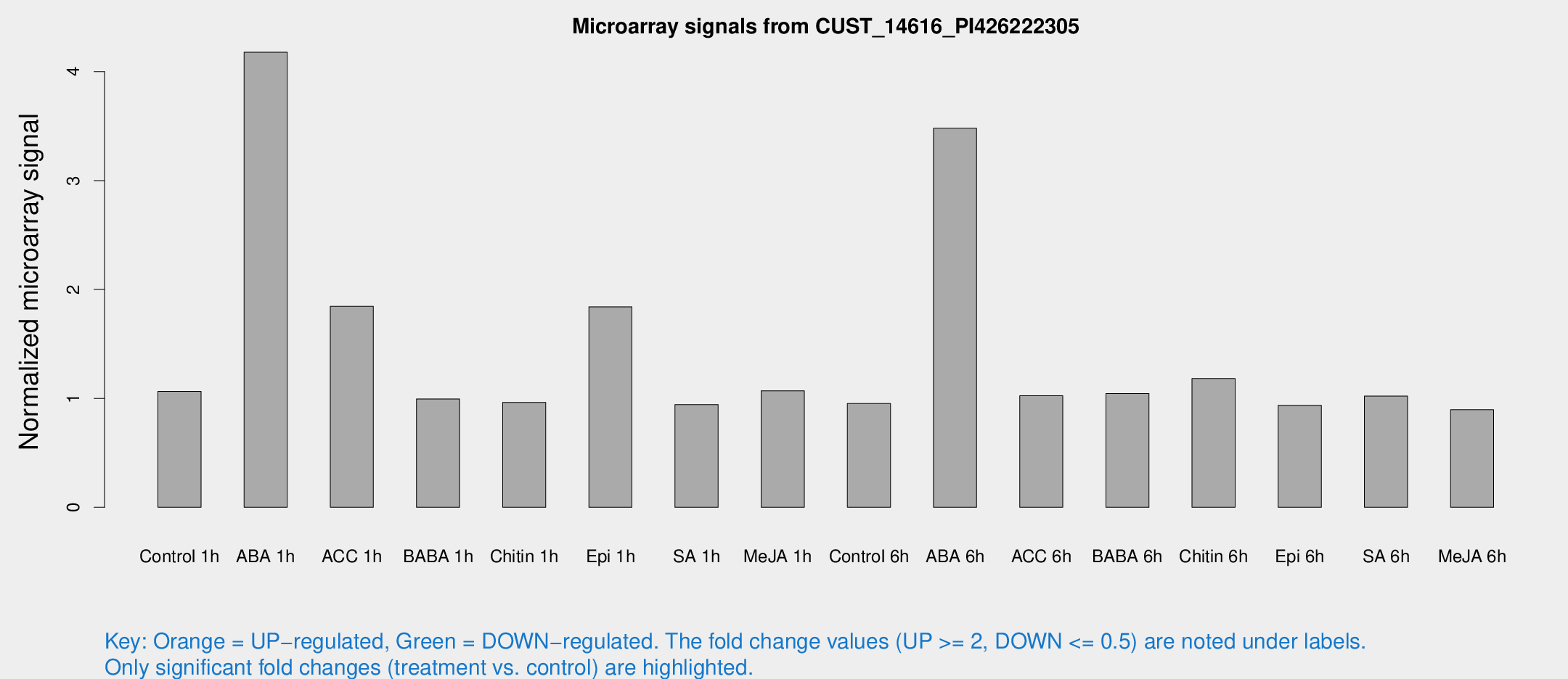
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.11088 | 3.13218 | 1.06429 | 0.521146 |
| ABA 1h | 29.6054 | 11.5482 | 4.17845 | 2.44927 |
| ACC 1h | 13.4984 | 3.95226 | 1.8451 | 0.849909 |
| BABA 1h | 6.09479 | 3.33499 | 0.995265 | 0.545045 |
| Chitin 1h | 5.46776 | 3.18027 | 0.963314 | 0.553167 |
| Epi 1h | 11.8831 | 4.90862 | 1.84121 | 0.744484 |
| SA 1h | 6.25309 | 3.15437 | 0.943097 | 0.49137 |
| Me-JA 1h | 5.5596 | 3.22629 | 1.0702 | 0.619886 |
| Control 6h | 6.08414 | 3.29669 | 0.952216 | 0.520008 |
| ABA 6h | 32.143 | 13.0759 | 3.48049 | 2.78831 |
| ACC 6h | 7.60115 | 4.02895 | 1.02413 | 0.533636 |
| BABA 6h | 7.72925 | 3.71017 | 1.04505 | 0.531528 |
| Chitin 6h | 8.50477 | 3.61906 | 1.18285 | 0.572905 |
| Epi 6h | 6.73119 | 3.78493 | 0.936423 | 0.521742 |
| SA 6h | 6.45421 | 3.45496 | 1.02129 | 0.55239 |
| Me-JA 6h | 5.66989 | 3.28595 | 0.895665 | 0.518643 |
Source Transcript PGSC0003DMT400066122 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G17870.1 | +1 | 5e-25 | 106 | 69/112 (62%) | ethylene-dependent gravitropism-deficient and yellow-green-like 3 | chr1:6150036-6152185 REVERSE LENGTH=573 |
