Probe CUST_14508_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14508_PI426222305 | JHI_St_60k_v1 | DMT400066573 | TCTTACAATGGATGGGCTACTTTGCTACAAGATATAACTTCATATACTTCCACAGGTCAT |
All Microarray Probes Designed to Gene DMG400025877
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14508_PI426222305 | JHI_St_60k_v1 | DMT400066573 | TCTTACAATGGATGGGCTACTTTGCTACAAGATATAACTTCATATACTTCCACAGGTCAT |
| CUST_14623_PI426222305 | JHI_St_60k_v1 | DMT400066574 | GCGTGTGTACGTTTTCTGTTTTGCTGATTTGATCATTACCTTTTGTCACCACTTCTAATA |
Microarray Signals from CUST_14508_PI426222305
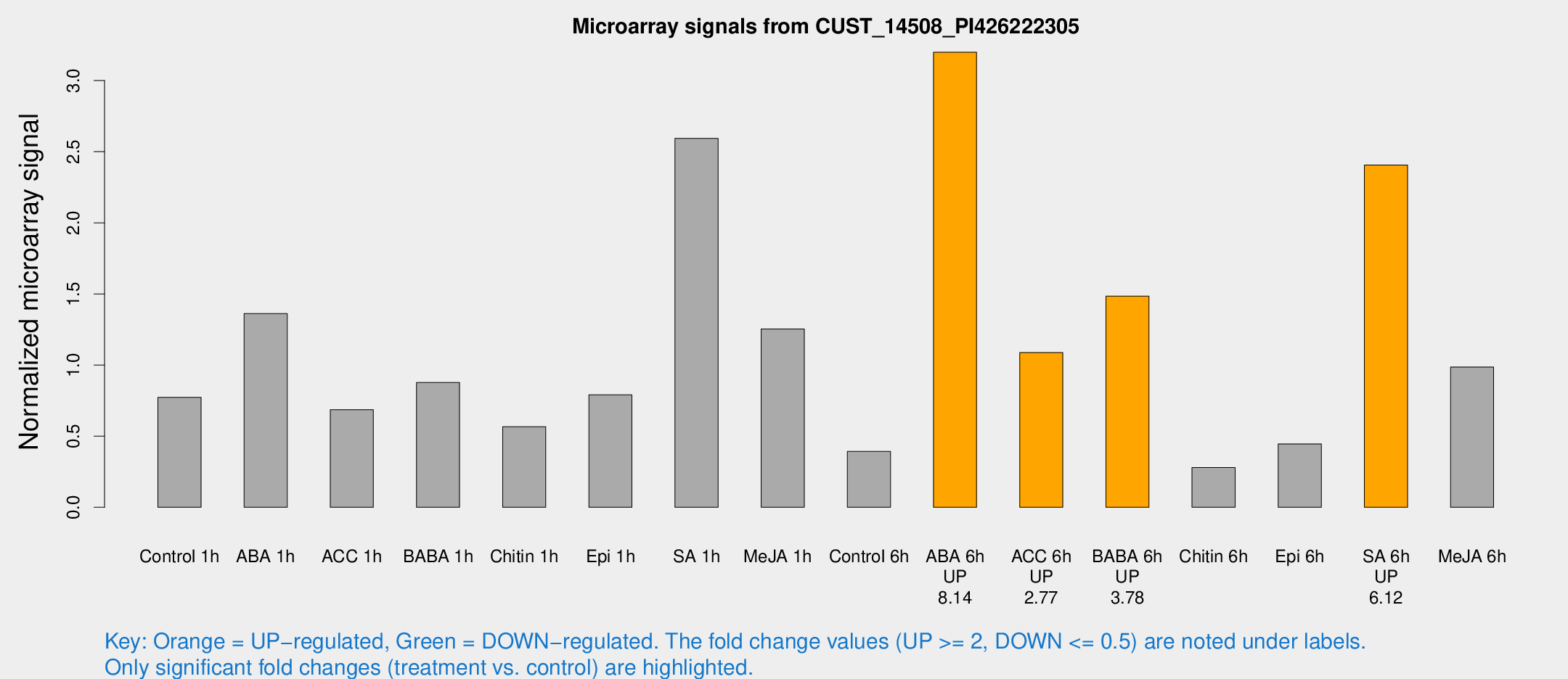
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4319.15 | 605.688 | 0.772653 | 0.0760914 |
| ABA 1h | 6792.75 | 1058.31 | 1.36145 | 0.173845 |
| ACC 1h | 4546.66 | 1552.67 | 0.686552 | 0.273182 |
| BABA 1h | 5108.87 | 1423.58 | 0.877289 | 0.204748 |
| Chitin 1h | 2894.13 | 552.031 | 0.566915 | 0.0781616 |
| Epi 1h | 3794.99 | 390.378 | 0.790752 | 0.0987991 |
| SA 1h | 15772.3 | 3986.34 | 2.59316 | 0.613151 |
| Me-JA 1h | 5853.62 | 1183.76 | 1.25426 | 0.146684 |
| Control 6h | 2219 | 340.292 | 0.392959 | 0.0319116 |
| ABA 6h | 20714.3 | 5782.84 | 3.19928 | 1.01351 |
| ACC 6h | 6991.44 | 1046.89 | 1.0879 | 0.285799 |
| BABA 6h | 9326.89 | 1326.74 | 1.48509 | 0.239215 |
| Chitin 6h | 1647.61 | 171.149 | 0.279001 | 0.026875 |
| Epi 6h | 2909.77 | 659.295 | 0.446004 | 0.0709526 |
| SA 6h | 13238.5 | 1576.36 | 2.40674 | 0.243447 |
| Me-JA 6h | 5729.92 | 1277.39 | 0.986434 | 0.308605 |
Source Transcript PGSC0003DMT400066573 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G34135.1 | +1 | 2e-65 | 211 | 104/175 (59%) | UDP-glucosyltransferase 73B2 | chr4:16345476-16347016 REVERSE LENGTH=483 |
