Probe CUST_13552_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13552_PI426222305 | JHI_St_60k_v1 | DMT400017405 | GCCACACGCACTGACTCTAGATTTTGGTTTCACCTCTGAATATAAACGTGCAGATTGTGA |
All Microarray Probes Designed to Gene DMG400006768
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13450_PI426222305 | JHI_St_60k_v1 | DMT400017406 | CCACCCATGCCTTCATGTTCGTGTGTTACATAACAATTTATGTAAATTGTAATTAGGTCA |
| CUST_13467_PI426222305 | JHI_St_60k_v1 | DMT400017404 | CATTTGGCTTGCATATGTTTTCTTATACTGTAGCAAAGCTGAAATCTTCCACGTTGCTAT |
| CUST_13545_PI426222305 | JHI_St_60k_v1 | DMT400017403 | ATCCGTCTATACGCTCATTATGCTTTGTAGATGGCAGAAAATAAGTGCTCCTGAGAGTAT |
| CUST_13552_PI426222305 | JHI_St_60k_v1 | DMT400017405 | GCCACACGCACTGACTCTAGATTTTGGTTTCACCTCTGAATATAAACGTGCAGATTGTGA |
Microarray Signals from CUST_13552_PI426222305
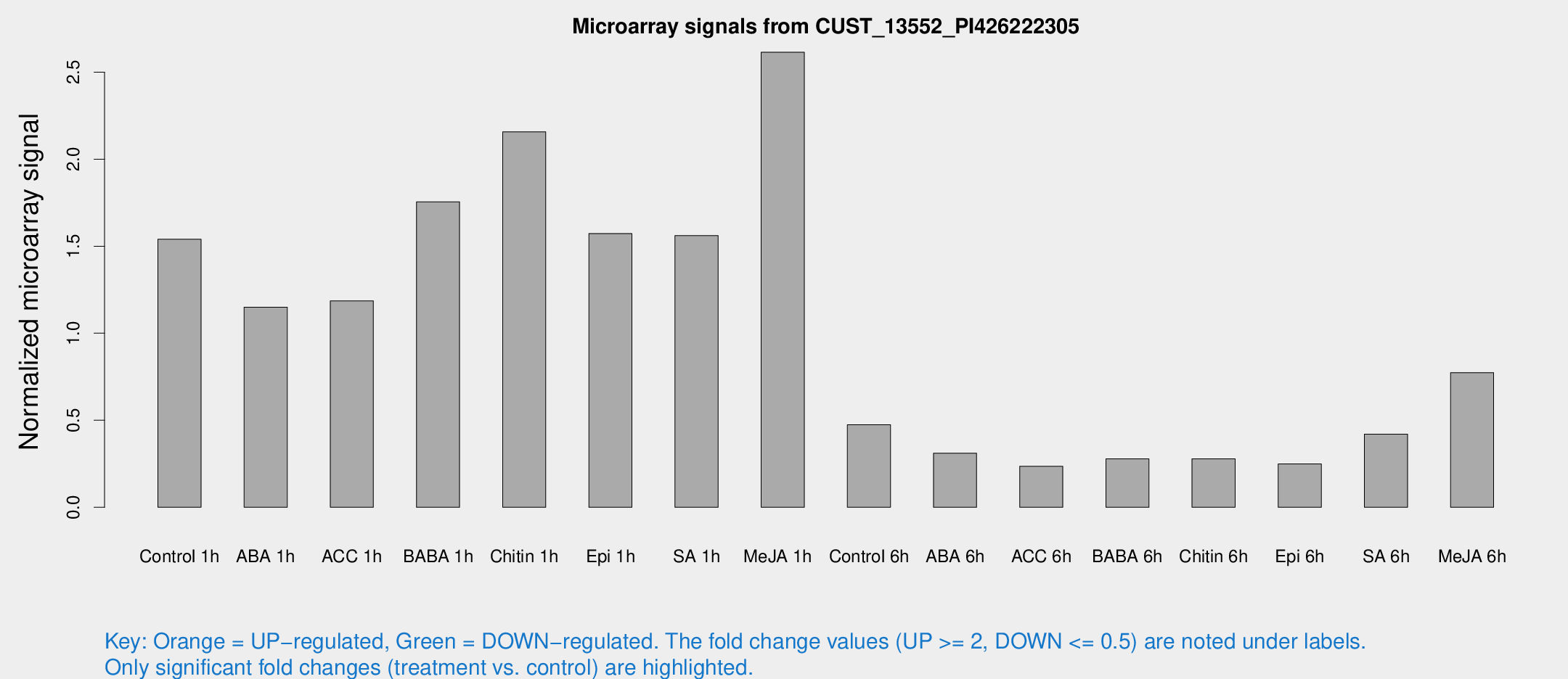
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 729.163 | 135.163 | 1.54008 | 0.178676 |
| ABA 1h | 468.91 | 36.696 | 1.14997 | 0.066874 |
| ACC 1h | 570.603 | 82.8357 | 1.18581 | 0.103465 |
| BABA 1h | 827.289 | 199.356 | 1.75498 | 0.299015 |
| Chitin 1h | 891.516 | 54.2686 | 2.15701 | 0.124799 |
| Epi 1h | 625.276 | 36.3663 | 1.57254 | 0.116035 |
| SA 1h | 740.158 | 63.7553 | 1.56084 | 0.090396 |
| Me-JA 1h | 993.018 | 126.369 | 2.6143 | 0.15122 |
| Control 6h | 228.675 | 46.1487 | 0.474045 | 0.0661498 |
| ABA 6h | 155.122 | 22.4814 | 0.311173 | 0.0343652 |
| ACC 6h | 132.515 | 36.0105 | 0.235531 | 0.087113 |
| BABA 6h | 144.313 | 15.6124 | 0.278333 | 0.0259954 |
| Chitin 6h | 138.855 | 21.1782 | 0.278689 | 0.0437638 |
| Epi 6h | 132.351 | 22.6365 | 0.24886 | 0.0548682 |
| SA 6h | 203.785 | 47.8611 | 0.420325 | 0.0674071 |
| Me-JA 6h | 360.528 | 51.6885 | 0.77381 | 0.0861206 |
Source Transcript PGSC0003DMT400017405 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G13800.2 | +3 | 1e-168 | 497 | 238/364 (65%) | pheophytinase | chr5:4452063-4454141 REVERSE LENGTH=484 |
