Probe CUST_13450_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13450_PI426222305 | JHI_St_60k_v1 | DMT400017406 | CCACCCATGCCTTCATGTTCGTGTGTTACATAACAATTTATGTAAATTGTAATTAGGTCA |
All Microarray Probes Designed to Gene DMG400006768
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_13450_PI426222305 | JHI_St_60k_v1 | DMT400017406 | CCACCCATGCCTTCATGTTCGTGTGTTACATAACAATTTATGTAAATTGTAATTAGGTCA |
| CUST_13467_PI426222305 | JHI_St_60k_v1 | DMT400017404 | CATTTGGCTTGCATATGTTTTCTTATACTGTAGCAAAGCTGAAATCTTCCACGTTGCTAT |
| CUST_13545_PI426222305 | JHI_St_60k_v1 | DMT400017403 | ATCCGTCTATACGCTCATTATGCTTTGTAGATGGCAGAAAATAAGTGCTCCTGAGAGTAT |
| CUST_13552_PI426222305 | JHI_St_60k_v1 | DMT400017405 | GCCACACGCACTGACTCTAGATTTTGGTTTCACCTCTGAATATAAACGTGCAGATTGTGA |
Microarray Signals from CUST_13450_PI426222305
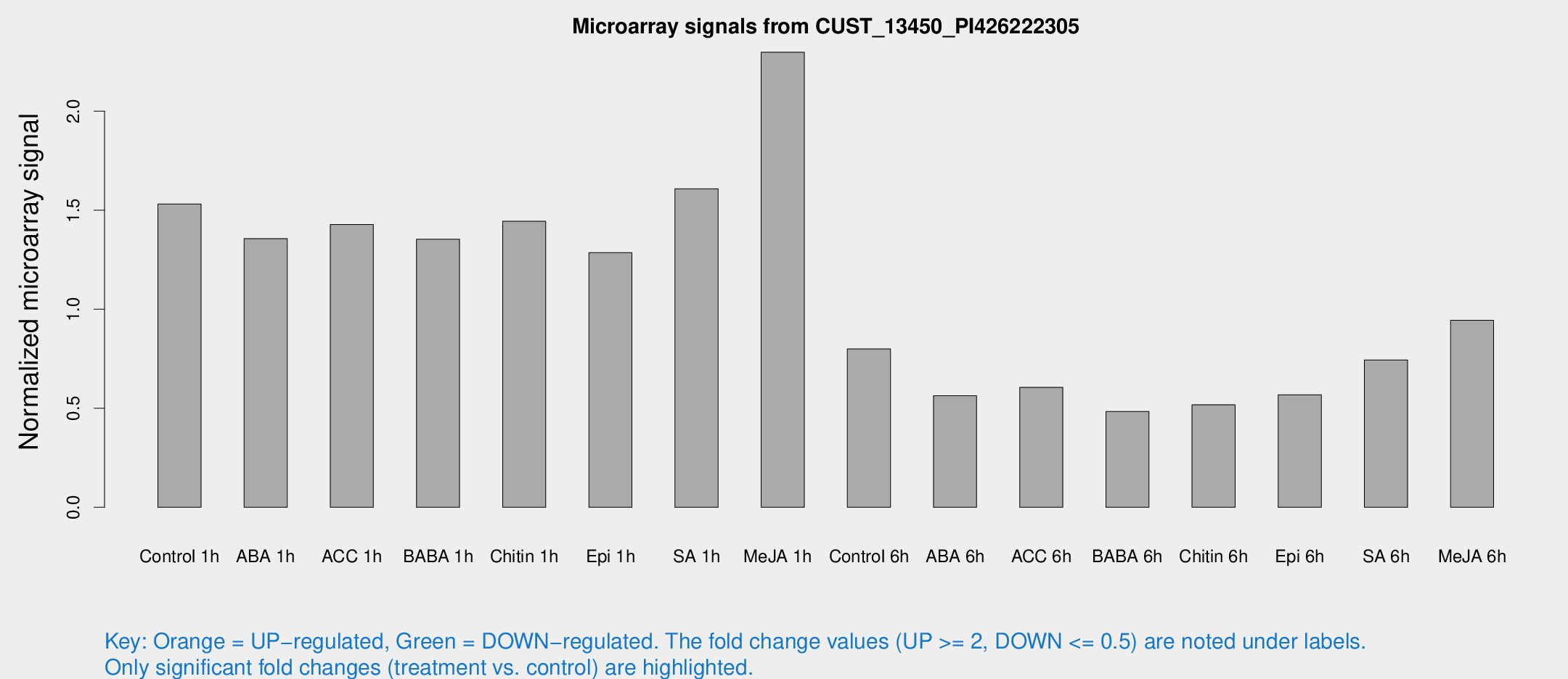
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 36210.1 | 2623.41 | 1.53111 | 0.0883988 |
| ABA 1h | 28569.3 | 3104.95 | 1.35701 | 0.0783471 |
| ACC 1h | 35201.9 | 4911.73 | 1.42785 | 0.123344 |
| BABA 1h | 33351.3 | 8573.02 | 1.35409 | 0.277631 |
| Chitin 1h | 30895 | 3252.98 | 1.44446 | 0.146328 |
| Epi 1h | 26317.8 | 1793.65 | 1.28633 | 0.0852329 |
| SA 1h | 39672.8 | 5810.71 | 1.60846 | 0.223392 |
| Me-JA 1h | 44650.1 | 4624.24 | 2.298 | 0.132675 |
| Control 6h | 19234.8 | 2745.47 | 0.799459 | 0.0552717 |
| ABA 6h | 14333.7 | 1840.91 | 0.563103 | 0.0462388 |
| ACC 6h | 16476.7 | 1404.42 | 0.60577 | 0.0845267 |
| BABA 6h | 12845.8 | 1075.83 | 0.484155 | 0.0604158 |
| Chitin 6h | 13288.8 | 2067.64 | 0.517878 | 0.0925064 |
| Epi 6h | 15426.9 | 2269.7 | 0.568514 | 0.118523 |
| SA 6h | 17787.2 | 2853.94 | 0.744167 | 0.055216 |
| Me-JA 6h | 22162.2 | 1281.8 | 0.944259 | 0.0549874 |
Source Transcript PGSC0003DMT400017406 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G13800.2 | +3 | 0.0 | 547 | 267/437 (61%) | pheophytinase | chr5:4452063-4454141 REVERSE LENGTH=484 |
