Probe CUST_12774_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12774_PI426222305 | JHI_St_60k_v1 | DMT400063026 | AATCTAGAGTTACAACCACAAGGGAGGAGCCTGATCATGCCTTCAAAAGGCGGAAAACAA |
All Microarray Probes Designed to Gene DMG400024523
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12720_PI426222305 | JHI_St_60k_v1 | DMT400063031 | CATGTGAAGGCTCAAGGACATTGTTCTAGTCTATTTTGTTATAGACATGATGAATTGCTA |
| CUST_12758_PI426222305 | JHI_St_60k_v1 | DMT400063029 | TCTACCTTGATACAGTACATTTATTTGCAGTTACTAACTGTATGATCACATTGCAGCCAA |
| CUST_12774_PI426222305 | JHI_St_60k_v1 | DMT400063026 | AATCTAGAGTTACAACCACAAGGGAGGAGCCTGATCATGCCTTCAAAAGGCGGAAAACAA |
| CUST_12789_PI426222305 | JHI_St_60k_v1 | DMT400063030 | AAGCAACGCTCATGTACTACTTCCACATGCATATCTATAGATGTGGTTGCAACAAATTAG |
Microarray Signals from CUST_12774_PI426222305
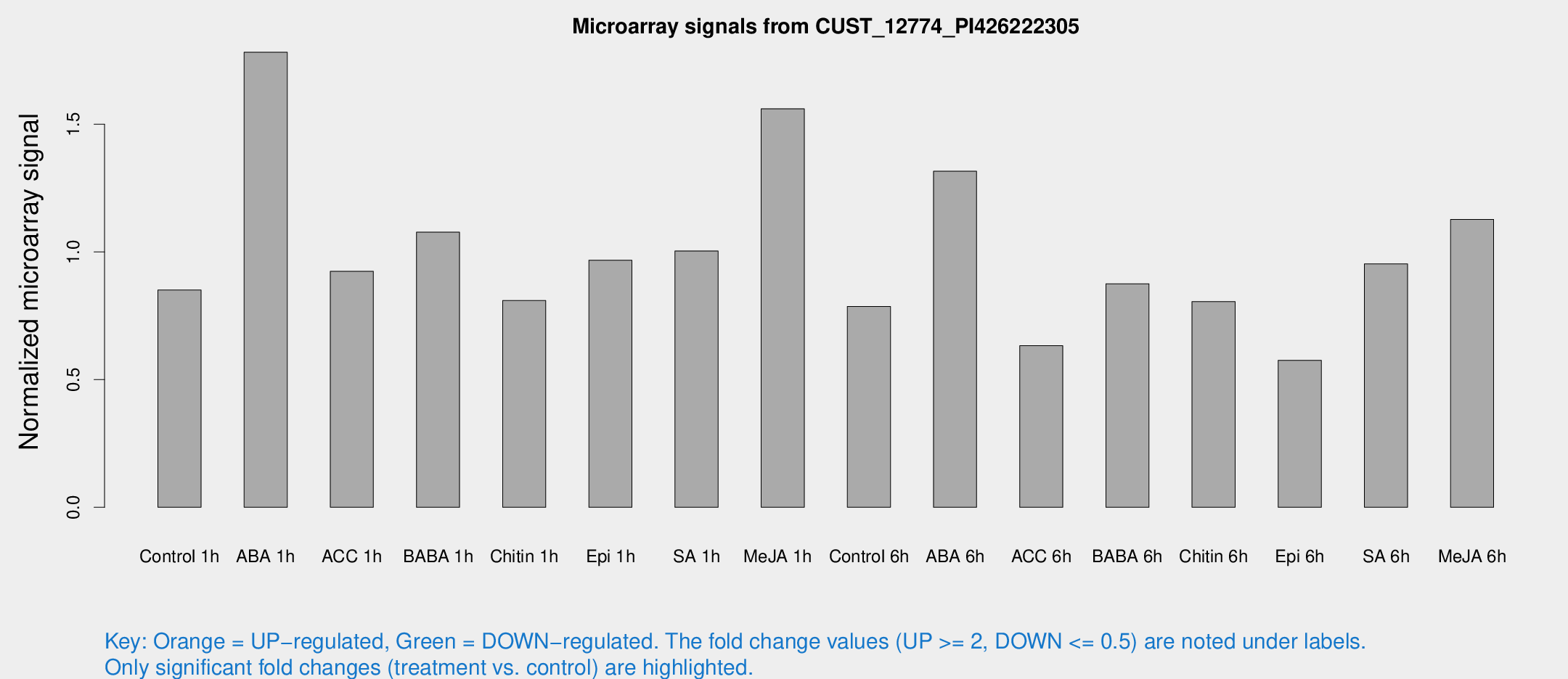
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 195.07 | 23.8526 | 0.850808 | 0.138706 |
| ABA 1h | 383.342 | 109.521 | 1.7821 | 0.367906 |
| ACC 1h | 223.086 | 43.3593 | 0.923918 | 0.132402 |
| BABA 1h | 241.783 | 42.6971 | 1.07736 | 0.110707 |
| Chitin 1h | 165.398 | 12.9671 | 0.809814 | 0.0942407 |
| Epi 1h | 204.235 | 49.9192 | 0.967719 | 0.288603 |
| SA 1h | 237.995 | 36.7202 | 1.00383 | 0.091308 |
| Me-JA 1h | 290.878 | 31.7765 | 1.56082 | 0.124692 |
| Control 6h | 197.052 | 58.377 | 0.78674 | 0.20822 |
| ABA 6h | 316.157 | 18.7135 | 1.31607 | 0.0777939 |
| ACC 6h | 167.943 | 27.6127 | 0.632688 | 0.0403088 |
| BABA 6h | 222.804 | 19.7071 | 0.875436 | 0.0696564 |
| Chitin 6h | 199.292 | 32.8845 | 0.80549 | 0.129141 |
| Epi 6h | 151.937 | 26.8625 | 0.575706 | 0.0797476 |
| SA 6h | 214.716 | 21.2545 | 0.953048 | 0.109314 |
| Me-JA 6h | 257.75 | 34.5849 | 1.12747 | 0.0674936 |
Source Transcript PGSC0003DMT400063026 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G08130.5 | +1 | 1e-76 | 254 | 226/544 (42%) | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | chr5:2606655-2609571 REVERSE LENGTH=532 |
