Probe CUST_12720_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12720_PI426222305 | JHI_St_60k_v1 | DMT400063031 | CATGTGAAGGCTCAAGGACATTGTTCTAGTCTATTTTGTTATAGACATGATGAATTGCTA |
All Microarray Probes Designed to Gene DMG400024523
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12720_PI426222305 | JHI_St_60k_v1 | DMT400063031 | CATGTGAAGGCTCAAGGACATTGTTCTAGTCTATTTTGTTATAGACATGATGAATTGCTA |
| CUST_12758_PI426222305 | JHI_St_60k_v1 | DMT400063029 | TCTACCTTGATACAGTACATTTATTTGCAGTTACTAACTGTATGATCACATTGCAGCCAA |
| CUST_12774_PI426222305 | JHI_St_60k_v1 | DMT400063026 | AATCTAGAGTTACAACCACAAGGGAGGAGCCTGATCATGCCTTCAAAAGGCGGAAAACAA |
| CUST_12789_PI426222305 | JHI_St_60k_v1 | DMT400063030 | AAGCAACGCTCATGTACTACTTCCACATGCATATCTATAGATGTGGTTGCAACAAATTAG |
Microarray Signals from CUST_12720_PI426222305
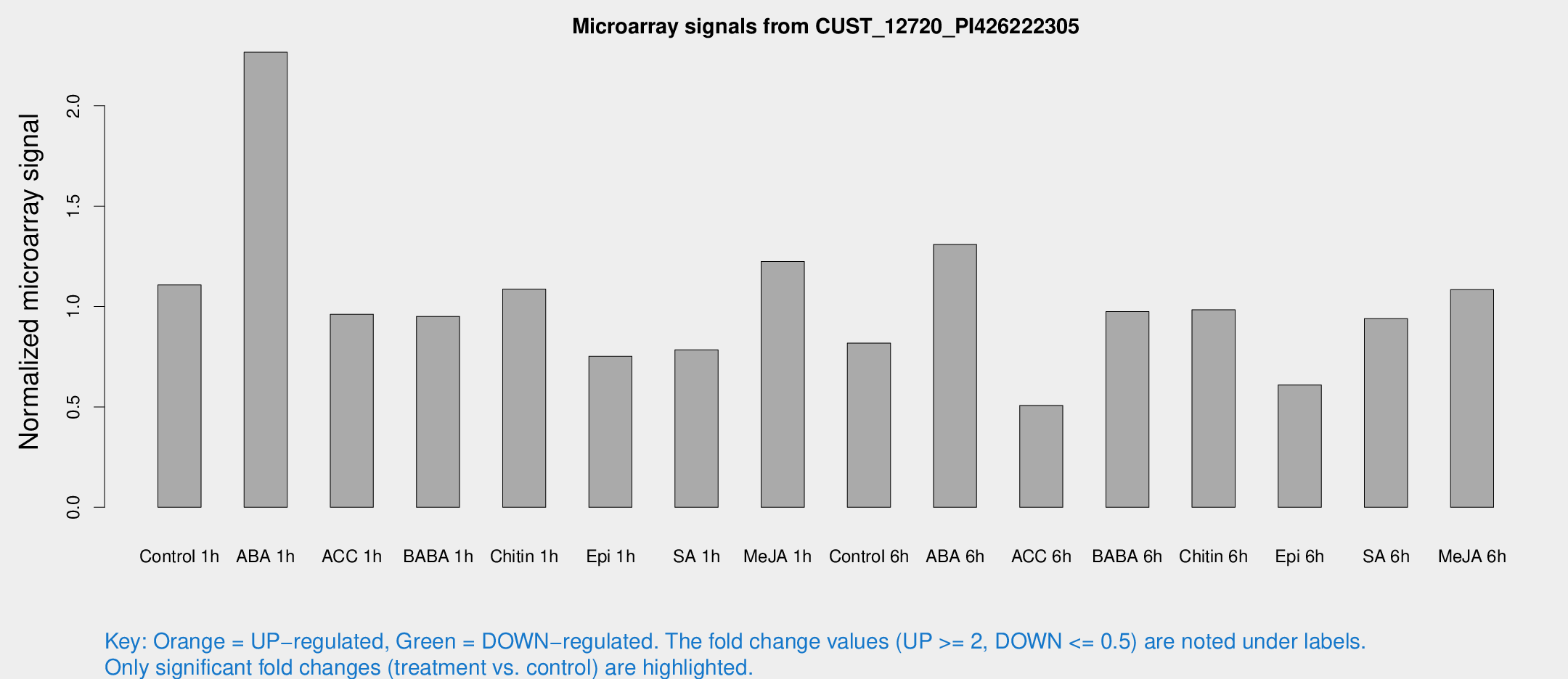
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 31.2233 | 7.34742 | 1.10785 | 0.210202 |
| ABA 1h | 53.8964 | 6.09265 | 2.26687 | 0.215198 |
| ACC 1h | 30.4317 | 11.0151 | 0.961324 | 0.339771 |
| BABA 1h | 25.2168 | 4.46886 | 0.951019 | 0.147687 |
| Chitin 1h | 25.9586 | 3.44748 | 1.08698 | 0.144397 |
| Epi 1h | 22.0079 | 8.39854 | 0.752201 | 0.513062 |
| SA 1h | 21.8489 | 3.33821 | 0.784457 | 0.124826 |
| Me-JA 1h | 29.0223 | 8.59584 | 1.224 | 0.344857 |
| Control 6h | 23.1598 | 5.18755 | 0.817418 | 0.146366 |
| ABA 6h | 38.4166 | 7.95736 | 1.30931 | 0.196965 |
| ACC 6h | 16.0133 | 4.00291 | 0.507218 | 0.131587 |
| BABA 6h | 32.9779 | 9.87993 | 0.974838 | 0.379917 |
| Chitin 6h | 28.4222 | 4.23815 | 0.983637 | 0.141906 |
| Epi 6h | 18.8097 | 3.83299 | 0.609328 | 0.132756 |
| SA 6h | 25.7269 | 5.48923 | 0.940036 | 0.324456 |
| Me-JA 6h | 29.8587 | 6.33098 | 1.08453 | 0.155044 |
Source Transcript PGSC0003DMT400063031 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G69010.1 | +3 | 1e-40 | 149 | 111/294 (38%) | BES1-interacting Myc-like protein 2 | chr1:25941804-25943599 FORWARD LENGTH=311 |
