Probe CUST_11355_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11355_PI426222305 | JHI_St_60k_v1 | DMT400010684 | TGTTGCTAATGGGAATTTGGACACCCCGATCATCAATAATGTGCCAAAGATGGTAGGCGT |
All Microarray Probes Designed to Gene DMG400004170
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11423_PI426222305 | JHI_St_60k_v1 | DMT400010685 | ATTTGGCTAGTTTCAACTTTCTGAGTCCATCTACATTGCTTTGTGACCCTATCCCCTCAC |
| CUST_11355_PI426222305 | JHI_St_60k_v1 | DMT400010684 | TGTTGCTAATGGGAATTTGGACACCCCGATCATCAATAATGTGCCAAAGATGGTAGGCGT |
Microarray Signals from CUST_11355_PI426222305
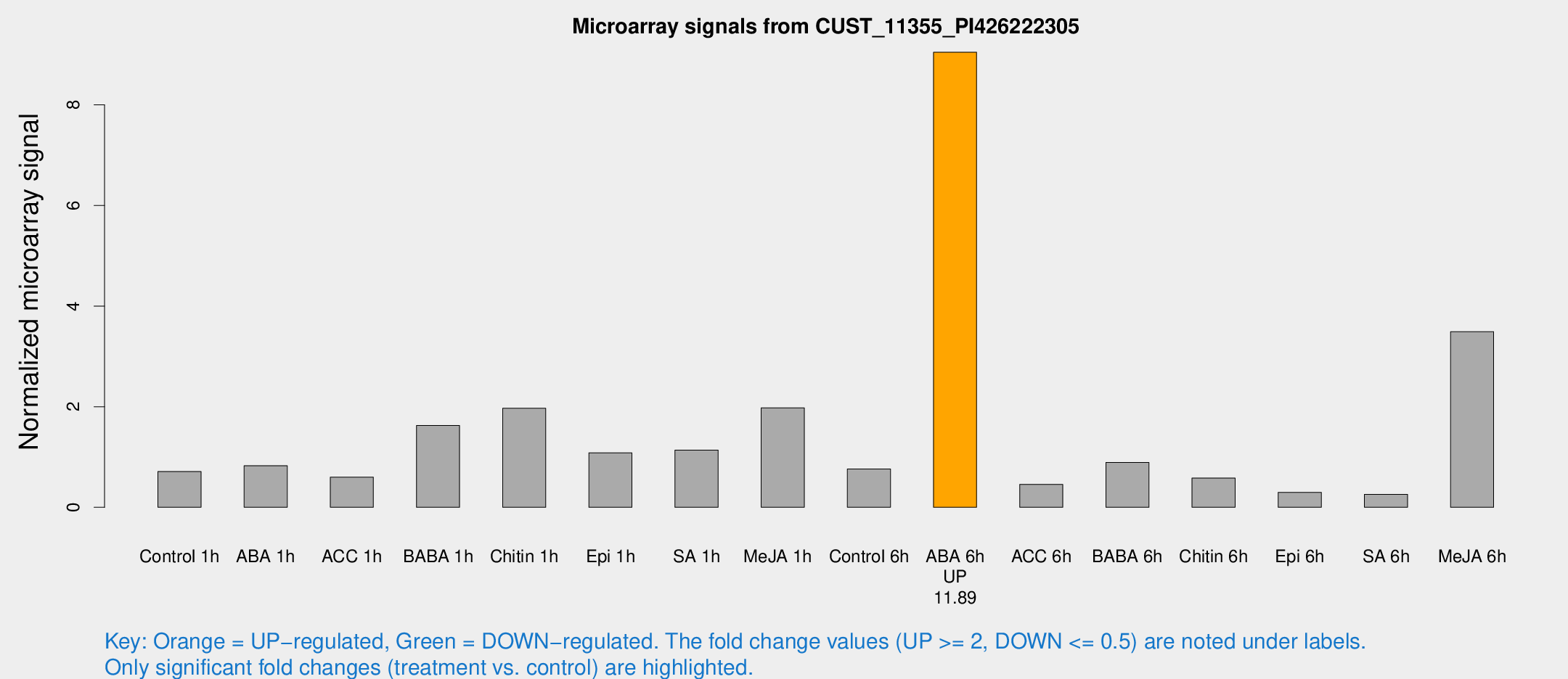
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 93.9223 | 10.9537 | 0.71025 | 0.136037 |
| ABA 1h | 132.5 | 57.8622 | 0.824991 | 0.651485 |
| ACC 1h | 92.1087 | 35.8356 | 0.598396 | 0.198475 |
| BABA 1h | 206.192 | 18.8619 | 1.62628 | 0.0981934 |
| Chitin 1h | 250.366 | 73.6757 | 1.96853 | 0.474801 |
| Epi 1h | 144.432 | 61.7435 | 1.08355 | 0.498423 |
| SA 1h | 155.687 | 22.6435 | 1.13861 | 0.150042 |
| Me-JA 1h | 212.987 | 23.3698 | 1.97833 | 0.279115 |
| Control 6h | 156.065 | 94.4148 | 0.761134 | 0.673312 |
| ABA 6h | 1297.3 | 237.42 | 9.04675 | 2.02698 |
| ACC 6h | 76.3585 | 21.8038 | 0.455724 | 0.163005 |
| BABA 6h | 133.193 | 19.4316 | 0.892105 | 0.111574 |
| Chitin 6h | 95.221 | 39.2144 | 0.580536 | 0.278464 |
| Epi 6h | 45.8294 | 9.9942 | 0.295274 | 0.0572575 |
| SA 6h | 33.6765 | 4.29575 | 0.256756 | 0.0339294 |
| Me-JA 6h | 502.294 | 169.542 | 3.49109 | 0.911441 |
Source Transcript PGSC0003DMT400010684 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G47340.1 | +1 | 0.0 | 859 | 423/512 (83%) | glutamine-dependent asparagine synthase 1 | chr3:17438136-17441043 REVERSE LENGTH=584 |
