Probe CUST_11423_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11423_PI426222305 | JHI_St_60k_v1 | DMT400010685 | ATTTGGCTAGTTTCAACTTTCTGAGTCCATCTACATTGCTTTGTGACCCTATCCCCTCAC |
All Microarray Probes Designed to Gene DMG400004170
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11423_PI426222305 | JHI_St_60k_v1 | DMT400010685 | ATTTGGCTAGTTTCAACTTTCTGAGTCCATCTACATTGCTTTGTGACCCTATCCCCTCAC |
| CUST_11355_PI426222305 | JHI_St_60k_v1 | DMT400010684 | TGTTGCTAATGGGAATTTGGACACCCCGATCATCAATAATGTGCCAAAGATGGTAGGCGT |
Microarray Signals from CUST_11423_PI426222305
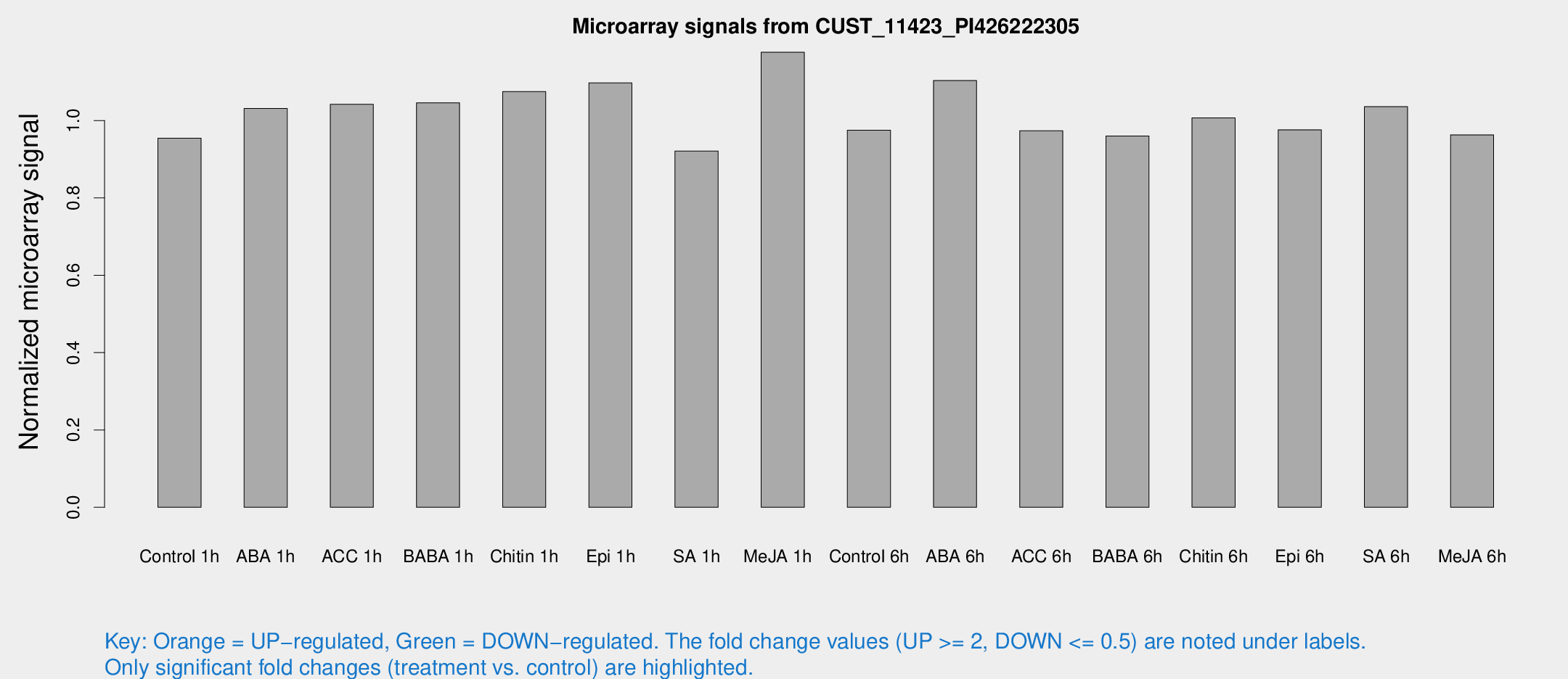
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.34259 | 3.09744 | 0.954322 | 0.552183 |
| ABA 1h | 5.07 | 2.93825 | 1.03142 | 0.593038 |
| ACC 1h | 6.01566 | 3.50006 | 1.04189 | 0.603331 |
| BABA 1h | 5.65109 | 3.27973 | 1.04595 | 0.606145 |
| Chitin 1h | 5.29218 | 3.08036 | 1.07505 | 0.606313 |
| Epi 1h | 5.23918 | 3.0405 | 1.09753 | 0.625087 |
| SA 1h | 5.29696 | 3.06839 | 0.921054 | 0.533373 |
| Me-JA 1h | 5.37921 | 3.13478 | 1.17667 | 0.6793 |
| Control 6h | 5.47424 | 3.17383 | 0.975191 | 0.564955 |
| ABA 6h | 6.67376 | 3.28579 | 1.10346 | 0.564871 |
| ACC 6h | 6.40063 | 3.80011 | 0.973594 | 0.563779 |
| BABA 6h | 6.02542 | 3.49498 | 0.960236 | 0.556054 |
| Chitin 6h | 6.00354 | 3.47832 | 1.00712 | 0.583148 |
| Epi 6h | 6.22318 | 3.64699 | 0.976157 | 0.565843 |
| SA 6h | 5.73507 | 3.32133 | 1.0359 | 0.599871 |
| Me-JA 6h | 5.3682 | 3.11017 | 0.963055 | 0.557875 |
Source Transcript PGSC0003DMT400010685 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G47340.1 | +2 | 0.0 | 993 | 488/585 (83%) | glutamine-dependent asparagine synthase 1 | chr3:17438136-17441043 REVERSE LENGTH=584 |
