Probe CUST_10867_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10867_PI426222305 | JHI_St_60k_v1 | DMT400031580 | GCAGTAAAATATCCTGGATCAGTCAAATCCCTCACTTTAATTGCACCGGTTTCCGAAGAA |
All Microarray Probes Designed to Gene DMG400012117
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10867_PI426222305 | JHI_St_60k_v1 | DMT400031580 | GCAGTAAAATATCCTGGATCAGTCAAATCCCTCACTTTAATTGCACCGGTTTCCGAAGAA |
| CUST_10634_PI426222305 | JHI_St_60k_v1 | DMT400031581 | GCAAGGGAGCTTGAACAAATTTGGAATAATTCAACATGTTCAAAATAATTTCCCACCATC |
| CUST_10702_PI426222305 | JHI_St_60k_v1 | DMT400031582 | AAACTGCCAACTATGCCTTAAAATTGGGAATTGATCAAGGGAAAAGCTATGTGAAGTAAA |
Microarray Signals from CUST_10867_PI426222305
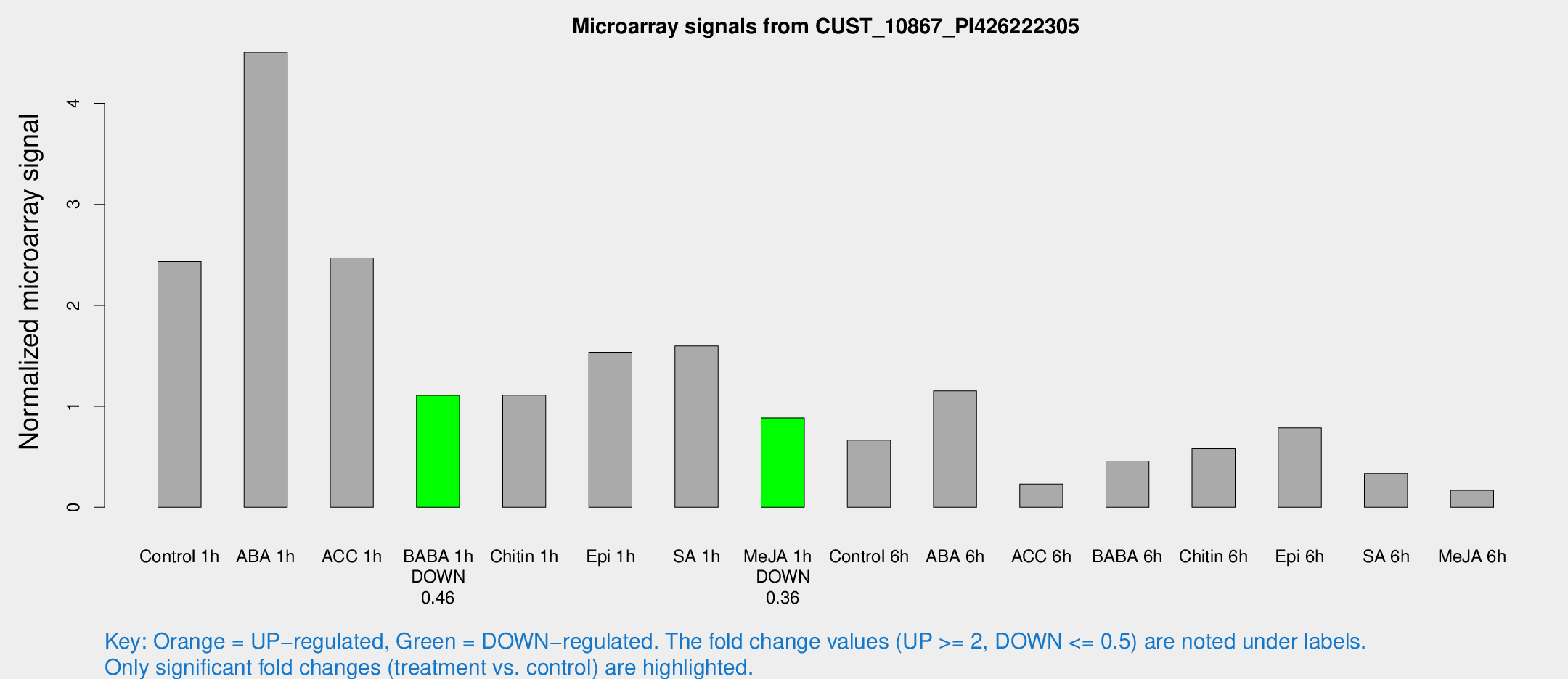
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 195.337 | 40.3535 | 2.4341 | 0.336157 |
| ABA 1h | 310.009 | 30.0942 | 4.50615 | 0.561237 |
| ACC 1h | 218.508 | 75.535 | 2.47044 | 0.702416 |
| BABA 1h | 82.313 | 5.87593 | 1.10898 | 0.117716 |
| Chitin 1h | 80.5608 | 18.4194 | 1.10972 | 0.262678 |
| Epi 1h | 104.378 | 13.9503 | 1.53632 | 0.201618 |
| SA 1h | 130.423 | 23.4675 | 1.59884 | 0.331675 |
| Me-JA 1h | 56.7208 | 7.45235 | 0.886353 | 0.183929 |
| Control 6h | 70.4538 | 33.9628 | 0.664738 | 0.441569 |
| ABA 6h | 96.2278 | 12.8899 | 1.15369 | 0.0992643 |
| ACC 6h | 20.4273 | 4.29215 | 0.230016 | 0.0469209 |
| BABA 6h | 42.3079 | 10.249 | 0.457876 | 0.112048 |
| Chitin 6h | 48.7588 | 7.39662 | 0.580657 | 0.107558 |
| Epi 6h | 69.0283 | 7.01919 | 0.786407 | 0.063315 |
| SA 6h | 26.3676 | 4.99319 | 0.334262 | 0.107451 |
| Me-JA 6h | 15.0151 | 4.81641 | 0.168032 | 0.0657842 |
Source Transcript PGSC0003DMT400031580 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G41900.1 | +1 | 2e-79 | 253 | 145/282 (51%) | alpha/beta-Hydrolases superfamily protein | chr5:16769032-16771567 FORWARD LENGTH=471 |
