Probe CUST_10634_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10634_PI426222305 | JHI_St_60k_v1 | DMT400031581 | GCAAGGGAGCTTGAACAAATTTGGAATAATTCAACATGTTCAAAATAATTTCCCACCATC |
All Microarray Probes Designed to Gene DMG400012117
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10867_PI426222305 | JHI_St_60k_v1 | DMT400031580 | GCAGTAAAATATCCTGGATCAGTCAAATCCCTCACTTTAATTGCACCGGTTTCCGAAGAA |
| CUST_10634_PI426222305 | JHI_St_60k_v1 | DMT400031581 | GCAAGGGAGCTTGAACAAATTTGGAATAATTCAACATGTTCAAAATAATTTCCCACCATC |
| CUST_10702_PI426222305 | JHI_St_60k_v1 | DMT400031582 | AAACTGCCAACTATGCCTTAAAATTGGGAATTGATCAAGGGAAAAGCTATGTGAAGTAAA |
Microarray Signals from CUST_10634_PI426222305
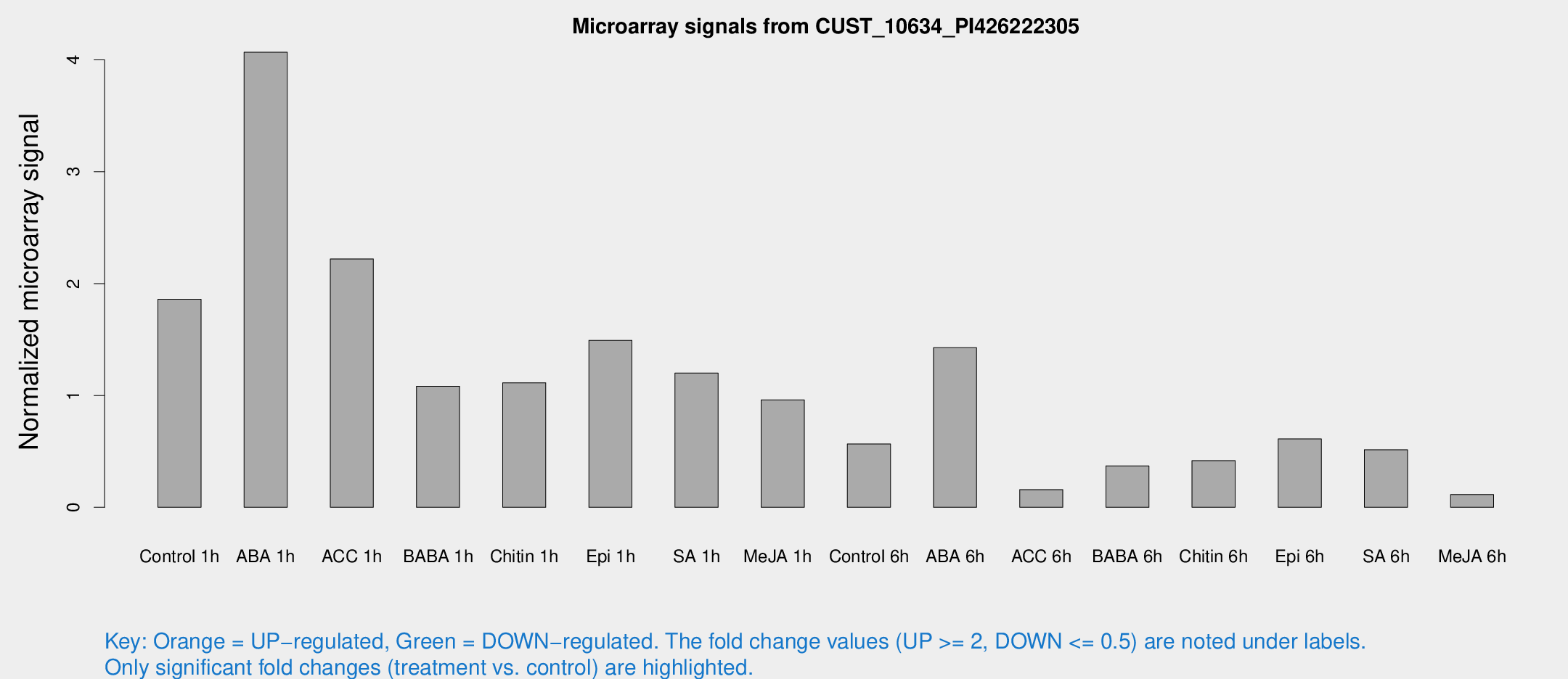
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 222.762 | 33.1788 | 1.85958 | 0.153586 |
| ABA 1h | 423.208 | 26.0202 | 4.06844 | 0.511491 |
| ACC 1h | 290.724 | 87.5348 | 2.22045 | 0.525313 |
| BABA 1h | 124.831 | 17.5445 | 1.08268 | 0.270037 |
| Chitin 1h | 124.975 | 33.0973 | 1.11332 | 0.299493 |
| Epi 1h | 152.236 | 11.5917 | 1.49278 | 0.0918312 |
| SA 1h | 149.681 | 27.6855 | 1.2005 | 0.283757 |
| Me-JA 1h | 93.6543 | 12.2785 | 0.960485 | 0.223325 |
| Control 6h | 82.4053 | 32.2401 | 0.566265 | 0.270686 |
| ABA 6h | 179.064 | 15.9129 | 1.42657 | 0.0871995 |
| ACC 6h | 22.3449 | 4.47832 | 0.158324 | 0.0320295 |
| BABA 6h | 49.7271 | 7.58936 | 0.370237 | 0.0430908 |
| Chitin 6h | 56.9798 | 16.549 | 0.417999 | 0.128405 |
| Epi 6h | 81.5321 | 7.9724 | 0.611013 | 0.0462665 |
| SA 6h | 64.8616 | 19.4094 | 0.515139 | 0.233324 |
| Me-JA 6h | 15.0994 | 4.56982 | 0.113556 | 0.0616173 |
Source Transcript PGSC0003DMT400031581 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G24140.1 | +3 | 8e-158 | 438 | 201/283 (71%) | alpha/beta-Hydrolases superfamily protein | chr4:12530032-12533664 REVERSE LENGTH=498 |
