Probe CUST_10818_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10818_PI426222305 | JHI_St_60k_v1 | DMT400032070 | CCATCAATTGTGATGGCAATAGTGAAATGCTTACTGTTTTGTCTCCCTCCACTTTGTAGA |
All Microarray Probes Designed to Gene DMG400012309
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10691_PI426222305 | JHI_St_60k_v1 | DMT400032065 | ATTTACTGCTATTGTTACATTGCTCCTTGTGACATCATGGTATACTAAATCGCTCTTCTT |
| CUST_10804_PI426222305 | JHI_St_60k_v1 | DMT400032067 | CACAGAGTGCACATTGTCCATTATTTAAGCATGGACTATTGATATCAATGTTATGGAGAT |
| CUST_10616_PI426222305 | JHI_St_60k_v1 | DMT400032069 | CACAGAGTGCACATTGTCCATTATTTAAGCATGGACTATTGATATCAATGTTATGGAGAT |
| CUST_10818_PI426222305 | JHI_St_60k_v1 | DMT400032070 | CCATCAATTGTGATGGCAATAGTGAAATGCTTACTGTTTTGTCTCCCTCCACTTTGTAGA |
Microarray Signals from CUST_10818_PI426222305
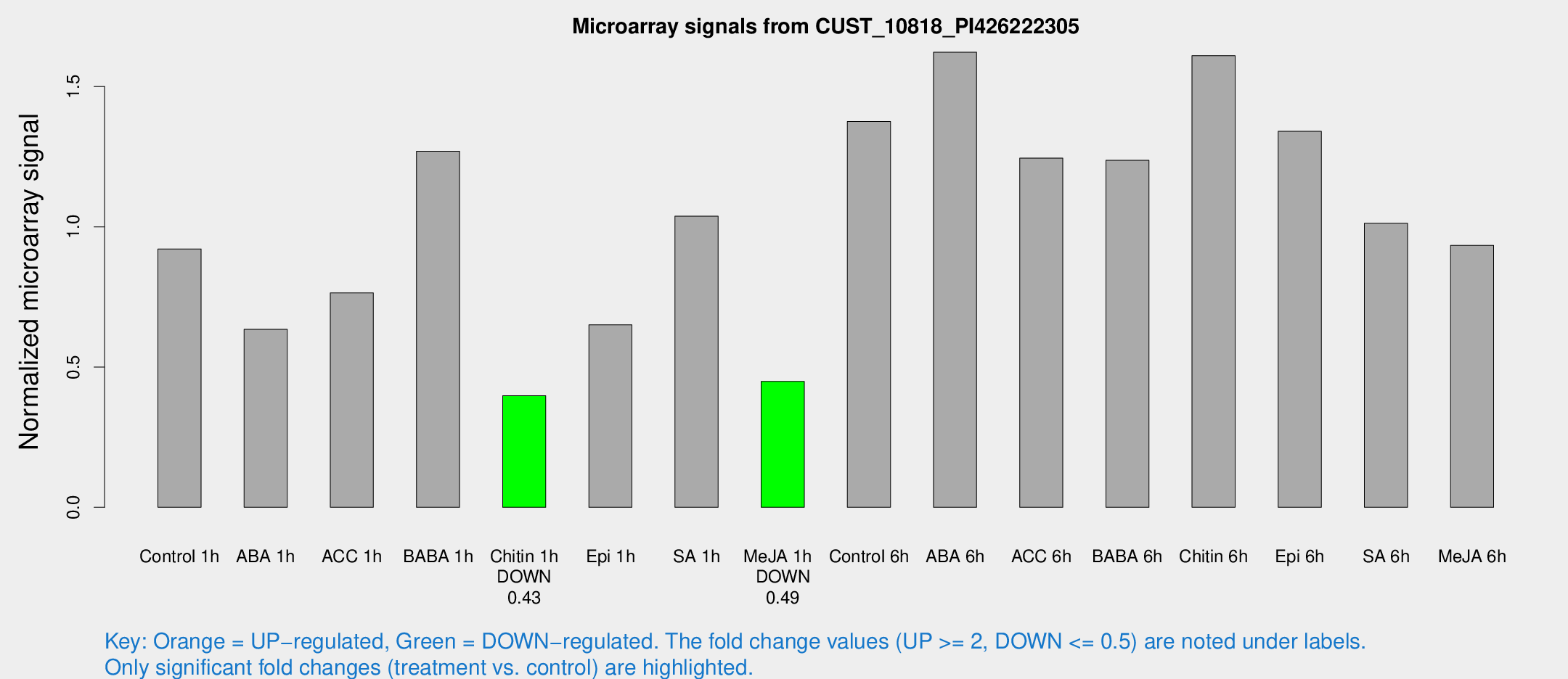
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 17.8828 | 3.60573 | 0.921127 | 0.191374 |
| ABA 1h | 11.6113 | 3.431 | 0.634665 | 0.214717 |
| ACC 1h | 15.9386 | 4.52972 | 0.764233 | 0.256461 |
| BABA 1h | 24.4671 | 5.14154 | 1.26943 | 0.220121 |
| Chitin 1h | 6.96189 | 3.46667 | 0.397842 | 0.201194 |
| Epi 1h | 15.5612 | 9.53315 | 0.650374 | 0.51451 |
| SA 1h | 20.7874 | 3.68911 | 1.03757 | 0.190134 |
| Me-JA 1h | 7.08708 | 3.60415 | 0.448749 | 0.23142 |
| Control 6h | 27.1896 | 4.47726 | 1.37515 | 0.210001 |
| ABA 6h | 36.1458 | 10.8957 | 1.62232 | 0.487543 |
| ACC 6h | 29.1549 | 7.06502 | 1.24481 | 0.398239 |
| BABA 6h | 26.7337 | 4.22387 | 1.23704 | 0.200659 |
| Chitin 6h | 35.3219 | 9.23371 | 1.61025 | 0.49331 |
| Epi 6h | 34.4056 | 12.611 | 1.34019 | 0.583636 |
| SA 6h | 27.2078 | 14.1992 | 1.01282 | 1.17533 |
| Me-JA 6h | 18.3923 | 3.71553 | 0.933532 | 0.203352 |
Source Transcript PGSC0003DMT400032070 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G53340.1 | +3 | 1e-17 | 82 | 34/43 (79%) | Galactosyltransferase family protein | chr5:21641045-21643195 REVERSE LENGTH=338 |
