Probe CUST_10616_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10616_PI426222305 | JHI_St_60k_v1 | DMT400032069 | CACAGAGTGCACATTGTCCATTATTTAAGCATGGACTATTGATATCAATGTTATGGAGAT |
All Microarray Probes Designed to Gene DMG400012309
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10691_PI426222305 | JHI_St_60k_v1 | DMT400032065 | ATTTACTGCTATTGTTACATTGCTCCTTGTGACATCATGGTATACTAAATCGCTCTTCTT |
| CUST_10804_PI426222305 | JHI_St_60k_v1 | DMT400032067 | CACAGAGTGCACATTGTCCATTATTTAAGCATGGACTATTGATATCAATGTTATGGAGAT |
| CUST_10616_PI426222305 | JHI_St_60k_v1 | DMT400032069 | CACAGAGTGCACATTGTCCATTATTTAAGCATGGACTATTGATATCAATGTTATGGAGAT |
| CUST_10818_PI426222305 | JHI_St_60k_v1 | DMT400032070 | CCATCAATTGTGATGGCAATAGTGAAATGCTTACTGTTTTGTCTCCCTCCACTTTGTAGA |
Microarray Signals from CUST_10616_PI426222305
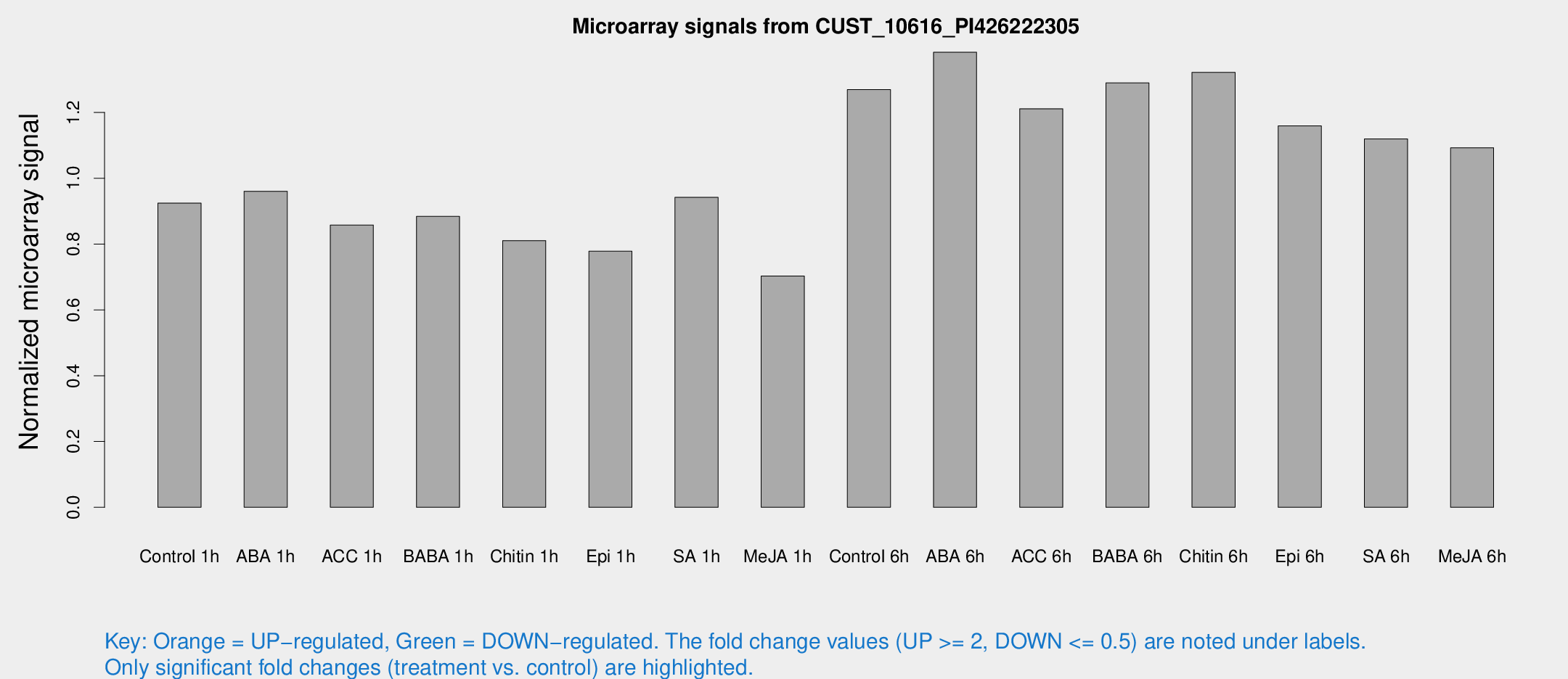
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 437.577 | 67.7397 | 0.924378 | 0.0809352 |
| ABA 1h | 393.193 | 22.9267 | 0.960697 | 0.0559693 |
| ACC 1h | 424.881 | 79.9262 | 0.857835 | 0.117442 |
| BABA 1h | 402.208 | 54.5097 | 0.884257 | 0.0516509 |
| Chitin 1h | 340.974 | 35.9226 | 0.810692 | 0.0647557 |
| Epi 1h | 316.474 | 38.0855 | 0.778399 | 0.0893494 |
| SA 1h | 447.912 | 26.0878 | 0.941982 | 0.0564854 |
| Me-JA 1h | 265.553 | 15.6883 | 0.703145 | 0.0656049 |
| Control 6h | 632.538 | 152.904 | 1.26959 | 0.243512 |
| ABA 6h | 688.887 | 76.6419 | 1.38323 | 0.0928441 |
| ACC 6h | 654.831 | 90.7638 | 1.21149 | 0.0703366 |
| BABA 6h | 671.372 | 49.3792 | 1.28999 | 0.0748129 |
| Chitin 6h | 652.428 | 37.9641 | 1.32195 | 0.0766978 |
| Epi 6h | 612.815 | 69.615 | 1.15916 | 0.222609 |
| SA 6h | 523.985 | 75.047 | 1.11983 | 0.0651594 |
| Me-JA 6h | 526.805 | 106.412 | 1.09282 | 0.151038 |
Source Transcript PGSC0003DMT400032069 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G53340.1 | +3 | 3e-97 | 305 | 135/200 (68%) | Galactosyltransferase family protein | chr5:21641045-21643195 REVERSE LENGTH=338 |
