Probe CUST_10783_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10783_PI426222305 | JHI_St_60k_v1 | DMT400031934 | CAGCATCTCAAAATCTGCACCCTAAGGGAATGTAATATTTATATAGACATCAATGGTTGA |
All Microarray Probes Designed to Gene DMG400012251
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10783_PI426222305 | JHI_St_60k_v1 | DMT400031934 | CAGCATCTCAAAATCTGCACCCTAAGGGAATGTAATATTTATATAGACATCAATGGTTGA |
| CUST_10658_PI426222305 | JHI_St_60k_v1 | DMT400031933 | TCAAAACTCTTGAAGACTATTCCCCAGGCAGCTGCAACAACTTGCTATGTTGCTACTCAC |
Microarray Signals from CUST_10783_PI426222305
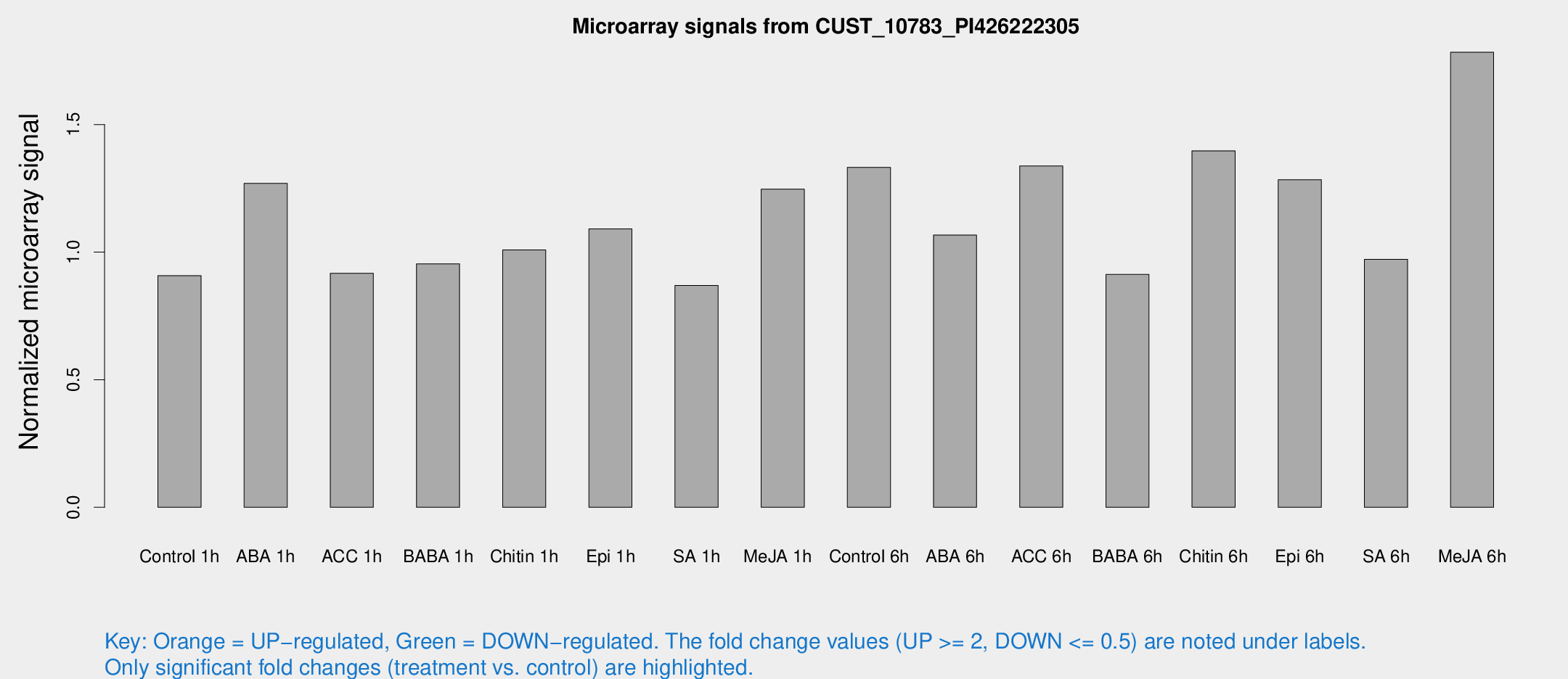
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.20129 | 2.88217 | 0.907611 | 0.503926 |
| ABA 1h | 6.73419 | 2.81266 | 1.26947 | 0.592965 |
| ACC 1h | 5.40016 | 3.13058 | 0.917169 | 0.531109 |
| BABA 1h | 5.27085 | 3.05849 | 0.954016 | 0.553159 |
| Chitin 1h | 5.05008 | 2.91736 | 1.0086 | 0.562908 |
| Epi 1h | 5.4221 | 2.90875 | 1.09141 | 0.588807 |
| SA 1h | 5.11213 | 2.91172 | 0.869482 | 0.495049 |
| Me-JA 1h | 5.87907 | 2.98082 | 1.24681 | 0.641059 |
| Control 6h | 8.18299 | 3.05329 | 1.33236 | 0.574387 |
| ABA 6h | 6.67475 | 3.16063 | 1.06678 | 0.528975 |
| ACC 6h | 9.53403 | 3.70515 | 1.33752 | 0.599205 |
| BABA 6h | 5.86301 | 3.4048 | 0.912589 | 0.528773 |
| Chitin 6h | 10.1492 | 4.45631 | 1.39699 | 0.647541 |
| Epi 6h | 9.29799 | 3.51225 | 1.28345 | 0.603726 |
| SA 6h | 5.50845 | 3.20543 | 0.971728 | 0.565085 |
| Me-JA 6h | 10.2842 | 3.08448 | 1.7833 | 0.554153 |
Source Transcript PGSC0003DMT400031934 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G24050.1 | +2 | 3e-148 | 436 | 253/383 (66%) | NAD(P)-binding Rossmann-fold superfamily protein | chr4:12497287-12499661 FORWARD LENGTH=332 |
