Probe CUST_10658_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10658_PI426222305 | JHI_St_60k_v1 | DMT400031933 | TCAAAACTCTTGAAGACTATTCCCCAGGCAGCTGCAACAACTTGCTATGTTGCTACTCAC |
All Microarray Probes Designed to Gene DMG400012251
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10783_PI426222305 | JHI_St_60k_v1 | DMT400031934 | CAGCATCTCAAAATCTGCACCCTAAGGGAATGTAATATTTATATAGACATCAATGGTTGA |
| CUST_10658_PI426222305 | JHI_St_60k_v1 | DMT400031933 | TCAAAACTCTTGAAGACTATTCCCCAGGCAGCTGCAACAACTTGCTATGTTGCTACTCAC |
Microarray Signals from CUST_10658_PI426222305
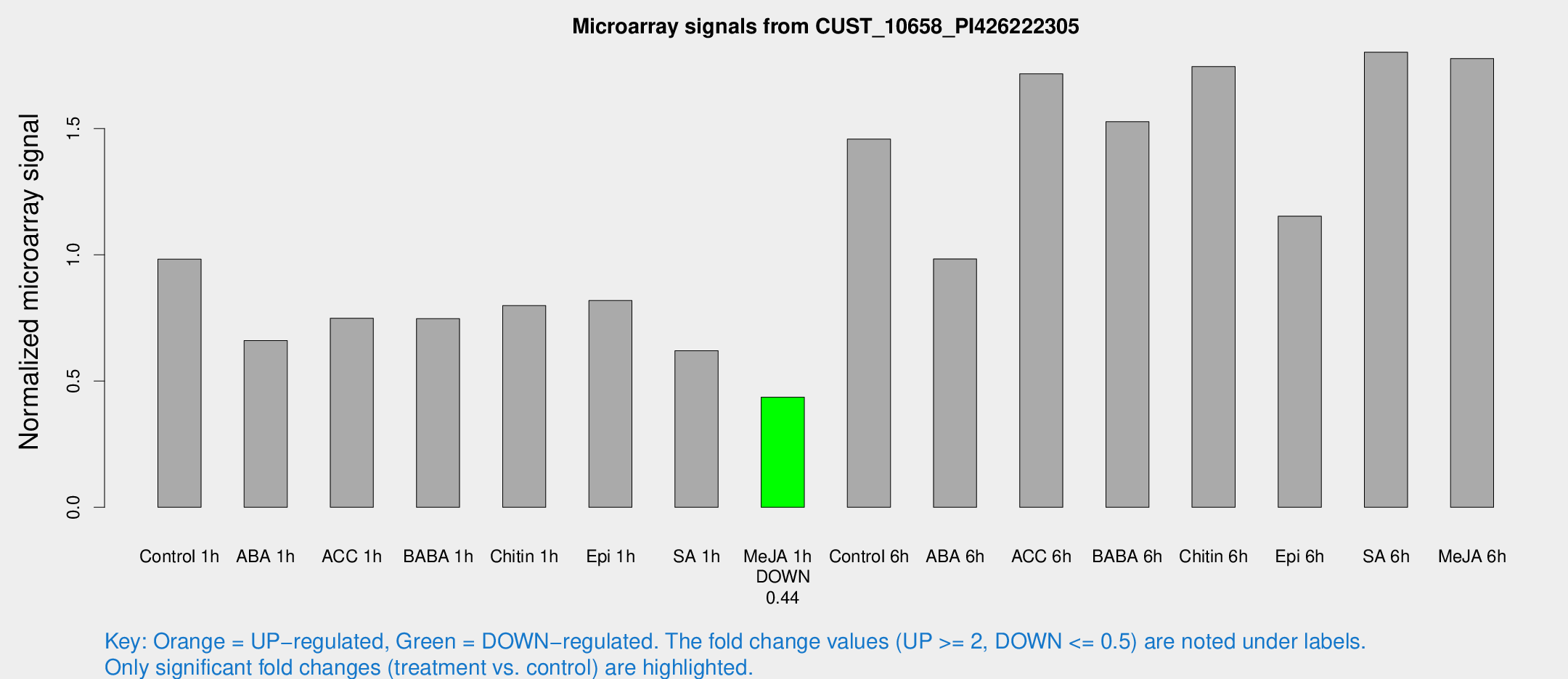
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 271.08 | 47.0671 | 0.982375 | 0.101902 |
| ABA 1h | 159.217 | 19.7393 | 0.660062 | 0.0519253 |
| ACC 1h | 222.084 | 55.3501 | 0.74888 | 0.158984 |
| BABA 1h | 204.876 | 47.1809 | 0.746822 | 0.117399 |
| Chitin 1h | 196.222 | 27.0688 | 0.798628 | 0.0476661 |
| Epi 1h | 194.626 | 30.8468 | 0.818741 | 0.10626 |
| SA 1h | 174.208 | 23.1484 | 0.620338 | 0.0537805 |
| Me-JA 1h | 96.3585 | 9.1933 | 0.436468 | 0.0286143 |
| Control 6h | 460.889 | 176.358 | 1.45818 | 0.544293 |
| ABA 6h | 280.639 | 16.5165 | 0.983604 | 0.0578303 |
| ACC 6h | 539.312 | 74.5534 | 1.71657 | 0.247601 |
| BABA 6h | 465.564 | 60.3137 | 1.52694 | 0.20036 |
| Chitin 6h | 526.601 | 130.187 | 1.74541 | 0.417785 |
| Epi 6h | 361.151 | 63.4576 | 1.15281 | 0.143123 |
| SA 6h | 515.458 | 131.964 | 1.80214 | 0.320543 |
| Me-JA 6h | 483.458 | 62.87 | 1.77707 | 0.118298 |
Source Transcript PGSC0003DMT400031933 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G24050.1 | +2 | 2e-158 | 460 | 253/331 (76%) | NAD(P)-binding Rossmann-fold superfamily protein | chr4:12497287-12499661 FORWARD LENGTH=332 |
