Probe CUST_10768_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10768_PI426222305 | JHI_St_60k_v1 | DMT400031777 | AGAAATTTGGGATGACTGATGAAATGCTAATTGACCAAGAATGGCAAAAGATAGCCAAGC |
All Microarray Probes Designed to Gene DMG400012191
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10842_PI426222305 | JHI_St_60k_v1 | DMT400031778 | TCGTGGAACATAACTGCAGCATTTGGTTCACTCCATATAGGGCATTAGGTATTCAACACA |
| CUST_10521_PI426222305 | JHI_St_60k_v1 | DMT400031779 | TCATGGATCTTGACGTTTTTTCGCGGTGCTTGTGAATGTATATGTAGGTACTGTTTGTCA |
| CUST_10768_PI426222305 | JHI_St_60k_v1 | DMT400031777 | AGAAATTTGGGATGACTGATGAAATGCTAATTGACCAAGAATGGCAAAAGATAGCCAAGC |
Microarray Signals from CUST_10768_PI426222305
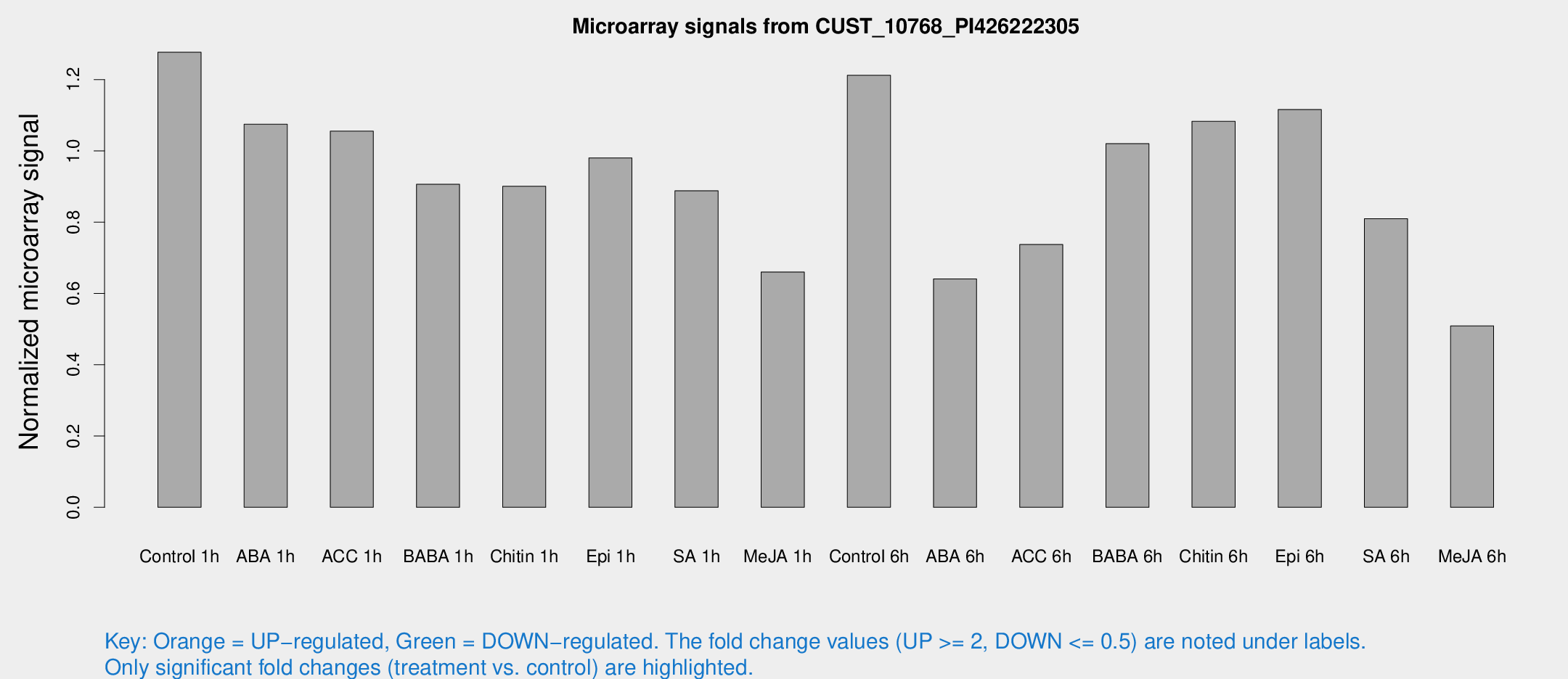
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2145.28 | 439.329 | 1.2769 | 0.177764 |
| ABA 1h | 1559.77 | 189.07 | 1.07489 | 0.100885 |
| ACC 1h | 1766.98 | 156.725 | 1.05539 | 0.060984 |
| BABA 1h | 1498.69 | 327.885 | 0.906455 | 0.137493 |
| Chitin 1h | 1319.56 | 114.426 | 0.900791 | 0.0520683 |
| Epi 1h | 1409.75 | 221.807 | 0.98044 | 0.146769 |
| SA 1h | 1522.43 | 254.713 | 0.888298 | 0.163698 |
| Me-JA 1h | 889.048 | 126.126 | 0.660272 | 0.038226 |
| Control 6h | 2187.38 | 660.443 | 1.21217 | 0.333438 |
| ABA 6h | 1103.9 | 63.981 | 0.640844 | 0.0388967 |
| ACC 6h | 1516.63 | 418.106 | 0.737372 | 0.210055 |
| BABA 6h | 1911.74 | 356.233 | 1.02038 | 0.170362 |
| Chitin 6h | 1880.41 | 182.103 | 1.08284 | 0.0709287 |
| Epi 6h | 2066.65 | 250.255 | 1.11624 | 0.205775 |
| SA 6h | 1439.47 | 398.525 | 0.809581 | 0.193747 |
| Me-JA 6h | 963.904 | 350.292 | 0.509063 | 0.188274 |
Source Transcript PGSC0003DMT400031777 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G05170.1 | +1 | 1e-91 | 287 | 141/264 (53%) | Phosphoglycerate mutase family protein | chr3:1466738-1468219 FORWARD LENGTH=316 |
