Probe CUST_10521_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10521_PI426222305 | JHI_St_60k_v1 | DMT400031779 | TCATGGATCTTGACGTTTTTTCGCGGTGCTTGTGAATGTATATGTAGGTACTGTTTGTCA |
All Microarray Probes Designed to Gene DMG400012191
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10842_PI426222305 | JHI_St_60k_v1 | DMT400031778 | TCGTGGAACATAACTGCAGCATTTGGTTCACTCCATATAGGGCATTAGGTATTCAACACA |
| CUST_10521_PI426222305 | JHI_St_60k_v1 | DMT400031779 | TCATGGATCTTGACGTTTTTTCGCGGTGCTTGTGAATGTATATGTAGGTACTGTTTGTCA |
| CUST_10768_PI426222305 | JHI_St_60k_v1 | DMT400031777 | AGAAATTTGGGATGACTGATGAAATGCTAATTGACCAAGAATGGCAAAAGATAGCCAAGC |
Microarray Signals from CUST_10521_PI426222305
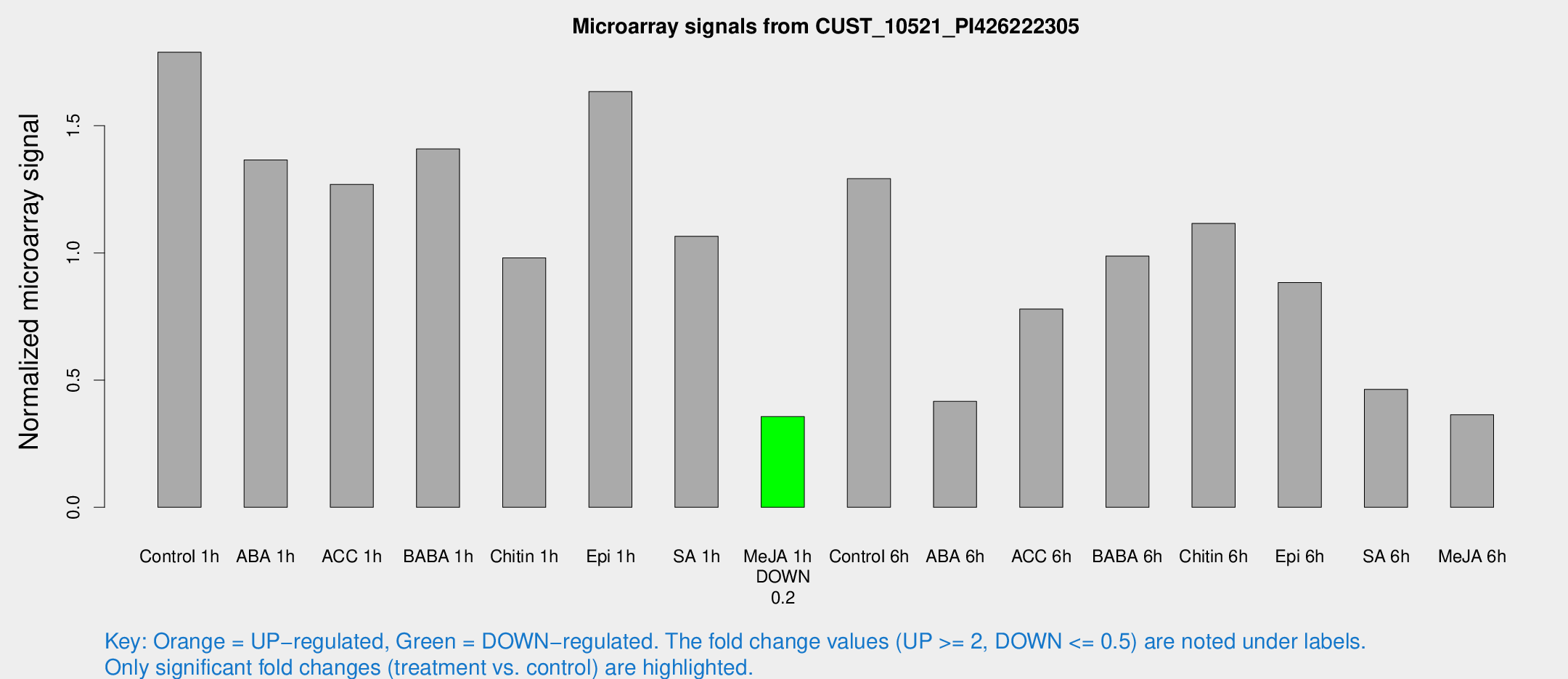
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 114.929 | 29.6276 | 1.78902 | 0.37085 |
| ABA 1h | 73.5308 | 6.528 | 1.36527 | 0.100728 |
| ACC 1h | 87.2149 | 28.7815 | 1.26957 | 0.339536 |
| BABA 1h | 89.372 | 23.8995 | 1.4091 | 0.299386 |
| Chitin 1h | 54.4196 | 7.55654 | 0.980854 | 0.088331 |
| Epi 1h | 91.3408 | 21.5017 | 1.63463 | 0.430163 |
| SA 1h | 67.6307 | 9.85206 | 1.06549 | 0.166439 |
| Me-JA 1h | 17.8809 | 3.55061 | 0.356753 | 0.0722012 |
| Control 6h | 88.8415 | 27.1889 | 1.29211 | 0.406276 |
| ABA 6h | 28.167 | 6.67426 | 0.41625 | 0.0806738 |
| ACC 6h | 55.0342 | 7.54598 | 0.779488 | 0.0754022 |
| BABA 6h | 81.3118 | 33.2036 | 0.987425 | 0.499568 |
| Chitin 6h | 74.4854 | 15.0715 | 1.11573 | 0.190688 |
| Epi 6h | 67.7327 | 21.145 | 0.883347 | 0.414784 |
| SA 6h | 33.7751 | 15.038 | 0.46331 | 0.181651 |
| Me-JA 6h | 32.4792 | 16.4832 | 0.363952 | 0.308149 |
Source Transcript PGSC0003DMT400031779 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G05170.1 | +1 | 1e-25 | 110 | 51/97 (53%) | Phosphoglycerate mutase family protein | chr3:1466738-1468219 FORWARD LENGTH=316 |
