Probe CUST_10601_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10601_PI426222305 | JHI_St_60k_v1 | DMT400031707 | AATGGGGGTATGTTGTCTTGCACGAAGAGGGATATCTATGAATTGTCATGTACGAGCCAG |
All Microarray Probes Designed to Gene DMG400012166
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_10601_PI426222305 | JHI_St_60k_v1 | DMT400031707 | AATGGGGGTATGTTGTCTTGCACGAAGAGGGATATCTATGAATTGTCATGTACGAGCCAG |
| CUST_10530_PI426222305 | JHI_St_60k_v1 | DMT400031712 | CTCAAAGTGGTGCTGTCAGAATAGAATGTTGATAGAATTTTCACTAAGAAATGAGCAATC |
| CUST_10542_PI426222305 | JHI_St_60k_v1 | DMT400031706 | AAATACACTTGACGCCAGATTAGAAGTTGTGTTCCGTAAGAAGCTACCAGAGGATGCTTC |
| CUST_10878_PI426222305 | JHI_St_60k_v1 | DMT400031711 | CTCAAAGTGGTGCTGTCAGAATAGAATGTTGATAGAATTTTCACTAAGAAATGAGCAATC |
Microarray Signals from CUST_10601_PI426222305
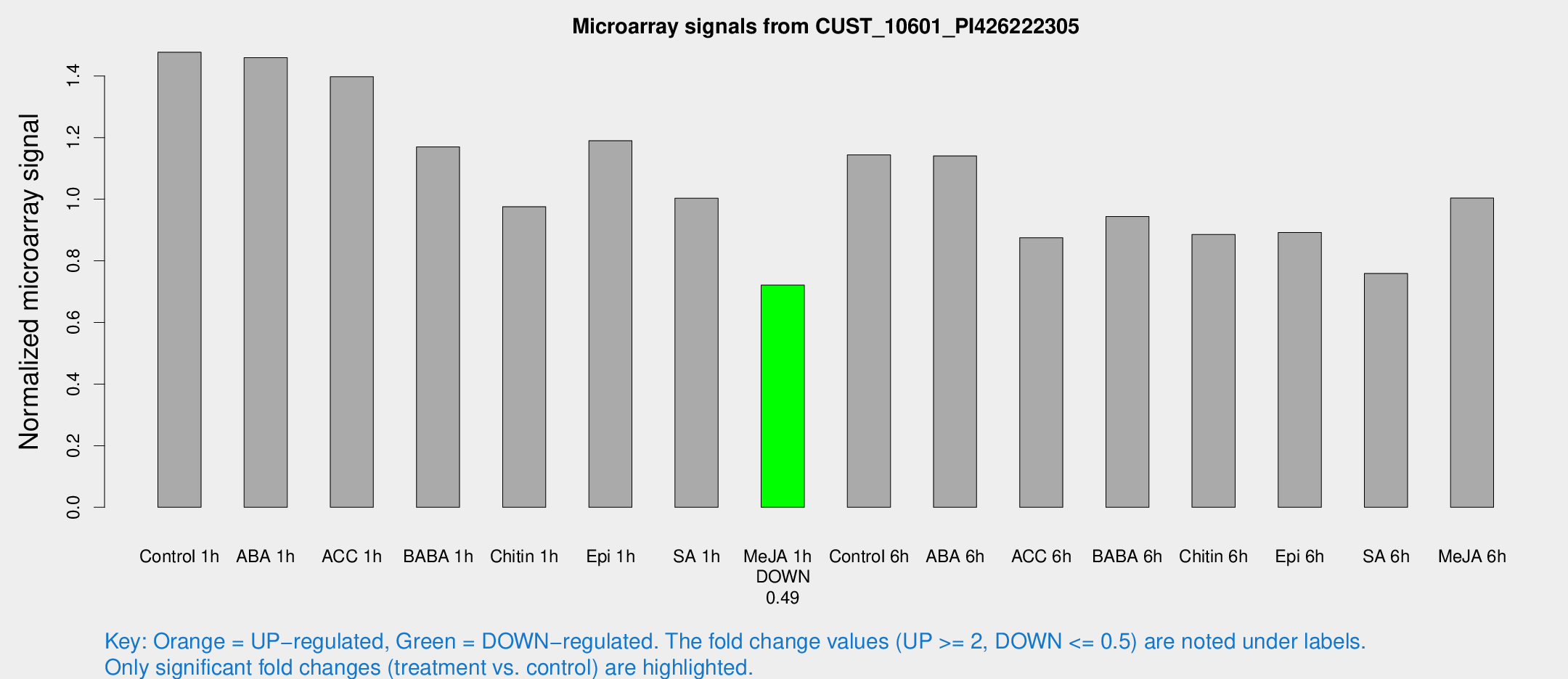
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 449.171 | 42.1607 | 1.47699 | 0.0889579 |
| ABA 1h | 389.915 | 22.7516 | 1.4591 | 0.156147 |
| ACC 1h | 453.209 | 94.2721 | 1.39727 | 0.221371 |
| BABA 1h | 348.44 | 53.4743 | 1.16968 | 0.0872336 |
| Chitin 1h | 266.518 | 22.2225 | 0.975672 | 0.0575065 |
| Epi 1h | 318.935 | 49.3083 | 1.19002 | 0.19042 |
| SA 1h | 333.202 | 89.4416 | 1.00293 | 0.256117 |
| Me-JA 1h | 178.317 | 11.6491 | 0.721087 | 0.0436099 |
| Control 6h | 372.36 | 90.3795 | 1.1439 | 0.231958 |
| ABA 6h | 370.428 | 40.9113 | 1.14047 | 0.0750365 |
| ACC 6h | 315.145 | 61.5887 | 0.875031 | 0.0882559 |
| BABA 6h | 319.741 | 18.8639 | 0.944002 | 0.0555328 |
| Chitin 6h | 284.639 | 16.8395 | 0.885657 | 0.0523444 |
| Epi 6h | 307.171 | 35.3857 | 0.892178 | 0.118969 |
| SA 6h | 227.27 | 14.1832 | 0.758916 | 0.10977 |
| Me-JA 6h | 303.93 | 28.0499 | 1.00354 | 0.0589327 |
Source Transcript PGSC0003DMT400031707 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G64200.1 | +2 | 7e-112 | 332 | 182/234 (78%) | vacuolar H+-ATPase subunit E isoform 3 | chr1:23828537-23830002 REVERSE LENGTH=237 |
