Probe CUST_1011_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1011_PI426222305 | JHI_St_60k_v1 | DMT400001357 | GTTGTTTCATTCACAAAGATTTTCATGATGTCGTTGACAGAATGAGTCTTCTCAACAAAC |
All Microarray Probes Designed to Gene DMG402000506
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1011_PI426222305 | JHI_St_60k_v1 | DMT400001357 | GTTGTTTCATTCACAAAGATTTTCATGATGTCGTTGACAGAATGAGTCTTCTCAACAAAC |
| CUST_1235_PI426222305 | JHI_St_60k_v1 | DMT400001358 | GTTGTTTCATTCACAAAGATTTTCATGATGTCGTTGACAGAATGAGTCTTCTCAACAAAC |
Microarray Signals from CUST_1011_PI426222305
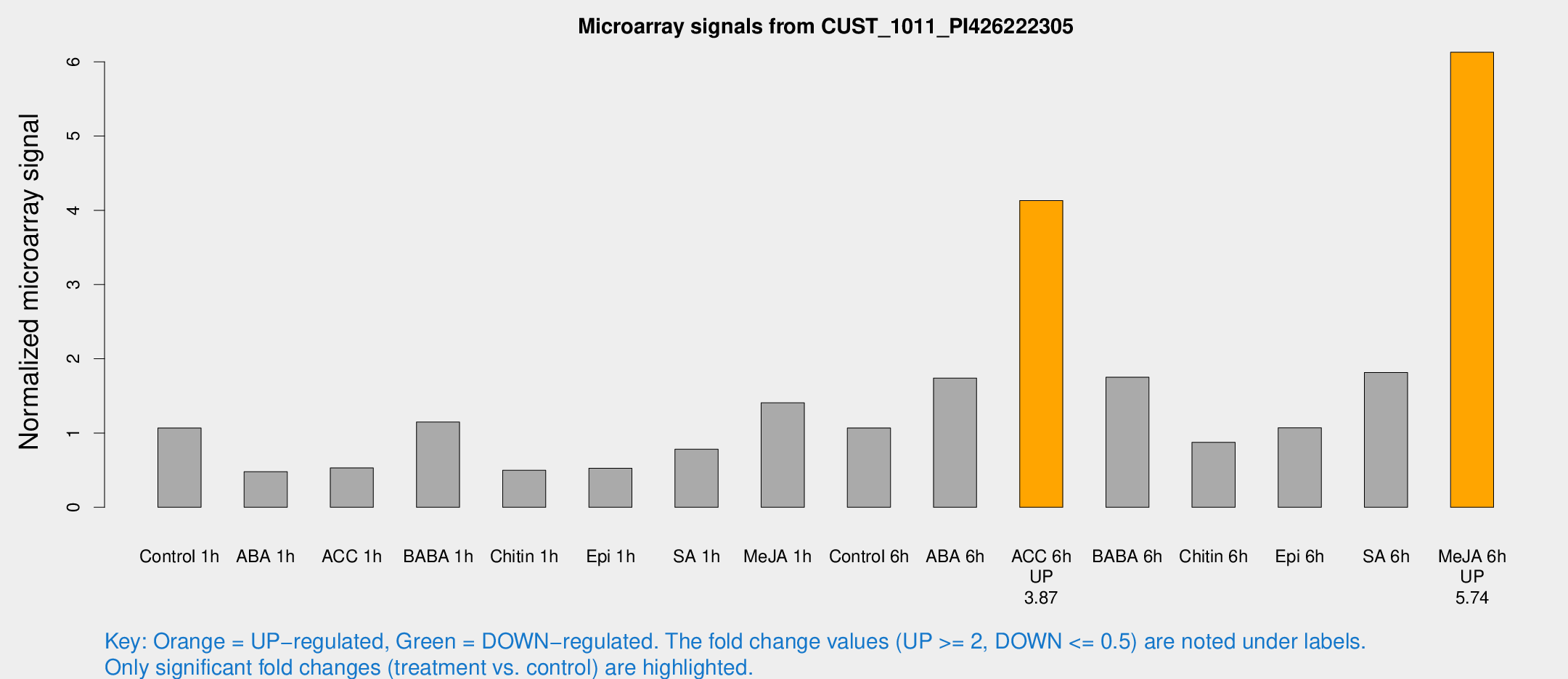
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13.3461 | 3.24557 | 1.06727 | 0.272666 |
| ABA 1h | 5.15228 | 2.98688 | 0.480034 | 0.276322 |
| ACC 1h | 6.6939 | 3.44606 | 0.529579 | 0.278137 |
| BABA 1h | 17.4156 | 8.38167 | 1.14793 | 0.570744 |
| Chitin 1h | 5.40182 | 3.14436 | 0.498253 | 0.284374 |
| Epi 1h | 5.5575 | 3.13026 | 0.525885 | 0.295387 |
| SA 1h | 10.72 | 3.17721 | 0.782574 | 0.287667 |
| Me-JA 1h | 16.9893 | 7.64081 | 1.40664 | 0.580061 |
| Control 6h | 14.2079 | 4.16317 | 1.06861 | 0.308806 |
| ABA 6h | 23.8094 | 5.08362 | 1.74103 | 0.33012 |
| ACC 6h | 62.2143 | 17.4355 | 4.13178 | 1.56844 |
| BABA 6h | 33.9605 | 19.7268 | 1.7514 | 1.2957 |
| Chitin 6h | 12.5822 | 3.63431 | 0.87522 | 0.320277 |
| Epi 6h | 14.7553 | 3.80158 | 1.07099 | 0.277746 |
| SA 6h | 22.6539 | 3.82293 | 1.81519 | 0.530857 |
| Me-JA 6h | 75.3296 | 7.90255 | 6.1289 | 0.442215 |
Source Transcript PGSC0003DMT400001357 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g087110.2 | +2 | 0.0 | 1196 | 593/656 (90%) | genomic_reference:SL2.50ch02 gene_region:44197070-44202789 transcript_region:SL2.50ch02:44197070..44202789+ go_terms:GO:0016165 functional_description:Alpha-dioxygenase (AHRD V1 **-- Q5GQ66_PEA); contains Interpro domain(s) IPR002007 Haem peroxidase, animal |
| TAIR PP10 | AT3G01420.1 | +2 | 0.0 | 910 | 447/646 (69%) | Peroxidase superfamily protein | chr3:159689-162726 REVERSE LENGTH=639 |
