Probe CUST_1235_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1235_PI426222305 | JHI_St_60k_v1 | DMT400001358 | GTTGTTTCATTCACAAAGATTTTCATGATGTCGTTGACAGAATGAGTCTTCTCAACAAAC |
All Microarray Probes Designed to Gene DMG402000506
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1011_PI426222305 | JHI_St_60k_v1 | DMT400001357 | GTTGTTTCATTCACAAAGATTTTCATGATGTCGTTGACAGAATGAGTCTTCTCAACAAAC |
| CUST_1235_PI426222305 | JHI_St_60k_v1 | DMT400001358 | GTTGTTTCATTCACAAAGATTTTCATGATGTCGTTGACAGAATGAGTCTTCTCAACAAAC |
Microarray Signals from CUST_1235_PI426222305
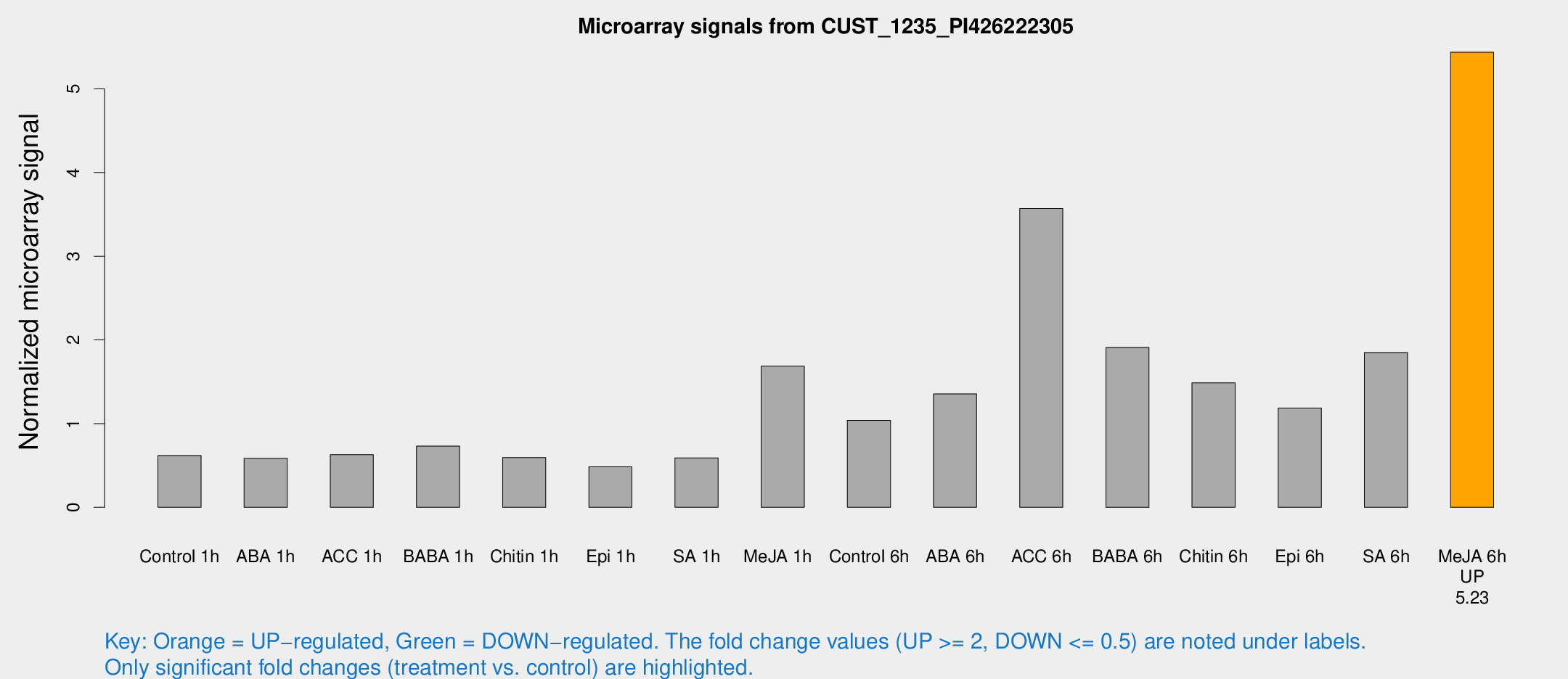
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.42744 | 4.257 | 0.618385 | 0.273587 |
| ABA 1h | 7.02779 | 2.79845 | 0.585299 | 0.274081 |
| ACC 1h | 9.43814 | 3.76223 | 0.628315 | 0.285011 |
| BABA 1h | 9.55473 | 3.09323 | 0.731212 | 0.279817 |
| Chitin 1h | 6.80213 | 2.92627 | 0.594889 | 0.257989 |
| Epi 1h | 5.18918 | 2.89759 | 0.483907 | 0.263794 |
| SA 1h | 8.10723 | 2.93407 | 0.590562 | 0.241864 |
| Me-JA 1h | 20.4877 | 8.00153 | 1.68609 | 0.692577 |
| Control 6h | 15.6397 | 5.17679 | 1.03907 | 0.415695 |
| ABA 6h | 21.8851 | 7.60189 | 1.35489 | 0.653875 |
| ACC 6h | 53.5045 | 9.67337 | 3.56997 | 0.953073 |
| BABA 6h | 41.1032 | 26.0151 | 1.91048 | 1.54637 |
| Chitin 6h | 20.5718 | 3.53115 | 1.48708 | 0.282033 |
| Epi 6h | 17.3975 | 3.60151 | 1.18696 | 0.252201 |
| SA 6h | 31.8757 | 13.3538 | 1.8495 | 2.13635 |
| Me-JA 6h | 69.4918 | 8.12113 | 5.43729 | 0.643243 |
Source Transcript PGSC0003DMT400001358 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g087110.2 | +2 | 0.0 | 1209 | 593/634 (94%) | genomic_reference:SL2.50ch02 gene_region:44197070-44202789 transcript_region:SL2.50ch02:44197070..44202789+ go_terms:GO:0016165 functional_description:Alpha-dioxygenase (AHRD V1 **-- Q5GQ66_PEA); contains Interpro domain(s) IPR002007 Haem peroxidase, animal |
| TAIR PP10 | AT3G01420.1 | +2 | 0.0 | 923 | 447/624 (72%) | Peroxidase superfamily protein | chr3:159689-162726 REVERSE LENGTH=639 |
