Probe CUST_49267_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49267_PI426222305 | JHI_St_60k_v1 | DMT400059101 | GCTTGTAGGTTGTATAGAAAAATTGAGGTGTTCATGGTAAAAGTGTCTGGATTTCTAGTT |
All Microarray Probes Designed to Gene DMG400022956
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49285_PI426222305 | JHI_St_60k_v1 | DMT400059103 | GCTTGTAGGTTGTATAGAAAAATTGAGGTGTTCATGGTAAAAGTGTCTGGATTTCTAGTT |
| CUST_49267_PI426222305 | JHI_St_60k_v1 | DMT400059101 | GCTTGTAGGTTGTATAGAAAAATTGAGGTGTTCATGGTAAAAGTGTCTGGATTTCTAGTT |
Microarray Signals from CUST_49267_PI426222305
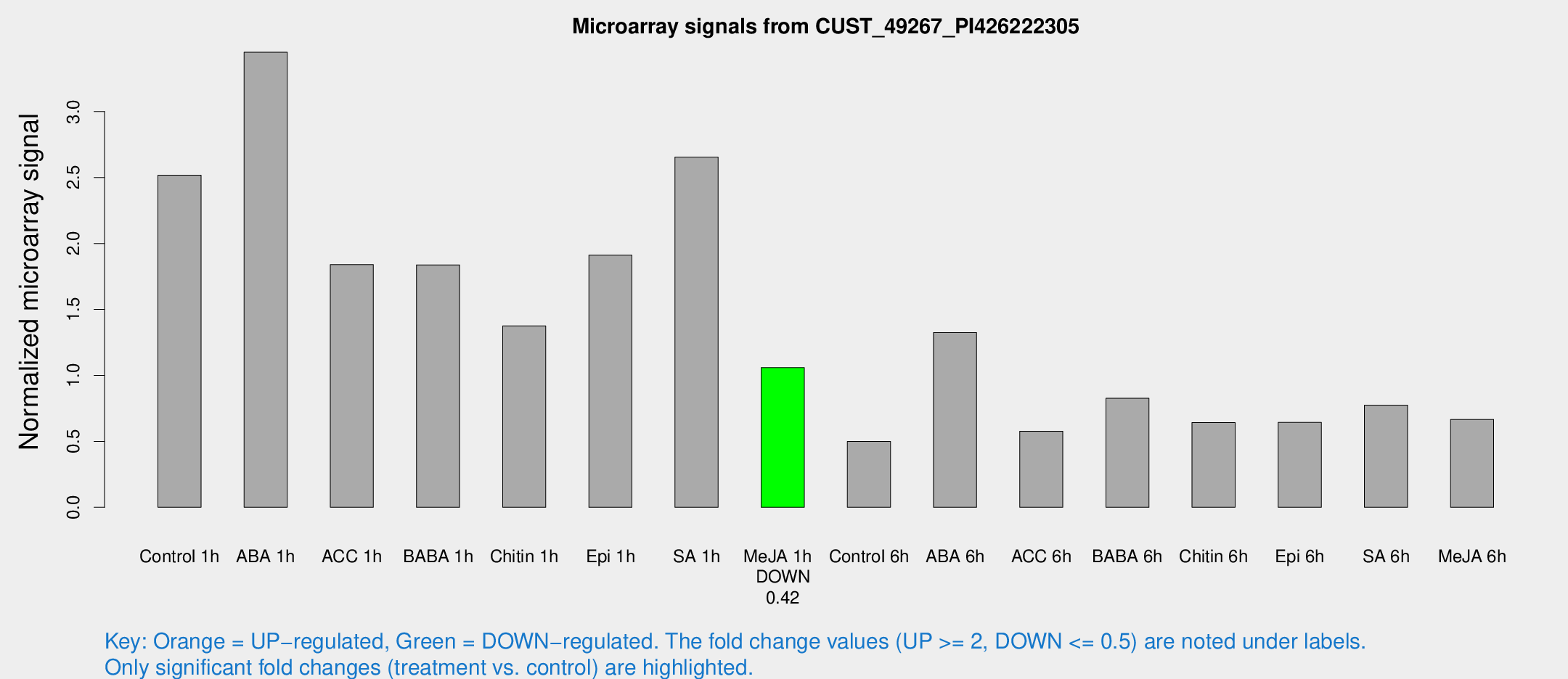
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3539.62 | 453.941 | 2.51742 | 0.217435 |
| ABA 1h | 4306.94 | 566.749 | 3.4501 | 0.299889 |
| ACC 1h | 2962.82 | 878.882 | 1.8396 | 0.601061 |
| BABA 1h | 2638.51 | 663.157 | 1.83802 | 0.341657 |
| Chitin 1h | 1721.07 | 129.784 | 1.37501 | 0.0885703 |
| Epi 1h | 2292.2 | 132.45 | 1.91213 | 0.11043 |
| SA 1h | 3991.89 | 989.264 | 2.65475 | 0.479478 |
| Me-JA 1h | 1230.54 | 214.015 | 1.05857 | 0.0930003 |
| Control 6h | 735.085 | 162.636 | 0.499652 | 0.0842773 |
| ABA 6h | 2041.5 | 409.325 | 1.32488 | 0.229129 |
| ACC 6h | 920.448 | 63.2853 | 0.576989 | 0.0409017 |
| BABA 6h | 1302.63 | 165.087 | 0.826387 | 0.0766765 |
| Chitin 6h | 947.219 | 54.8604 | 0.642314 | 0.0371734 |
| Epi 6h | 1024.94 | 135.631 | 0.643915 | 0.0525295 |
| SA 6h | 1099.01 | 191.478 | 0.774463 | 0.0589867 |
| Me-JA 6h | 979.46 | 251.317 | 0.665805 | 0.116592 |
Source Transcript PGSC0003DMT400059101 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G05190.1 | +1 | 4e-20 | 96 | 44/87 (51%) | Protein of unknown function (DUF3133) | chr5:1541853-1543875 FORWARD LENGTH=615 |
