Probe CUST_45797_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45797_PI426222305 | JHI_St_60k_v1 | DMT400050270 | TAACCTGCCGGTGTGGATCTCAATTTTGCTATATATGTGGAGAAACATGGAGCGAAAGCC |
All Microarray Probes Designed to Gene DMG400019526
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45797_PI426222305 | JHI_St_60k_v1 | DMT400050270 | TAACCTGCCGGTGTGGATCTCAATTTTGCTATATATGTGGAGAAACATGGAGCGAAAGCC |
| CUST_45809_PI426222305 | JHI_St_60k_v1 | DMT400050269 | CACATAACCTGCCGGTTAGTTAATTTCTTGTTTAATTTTTATATGGTAGTGTGGCCTGTT |
Microarray Signals from CUST_45797_PI426222305
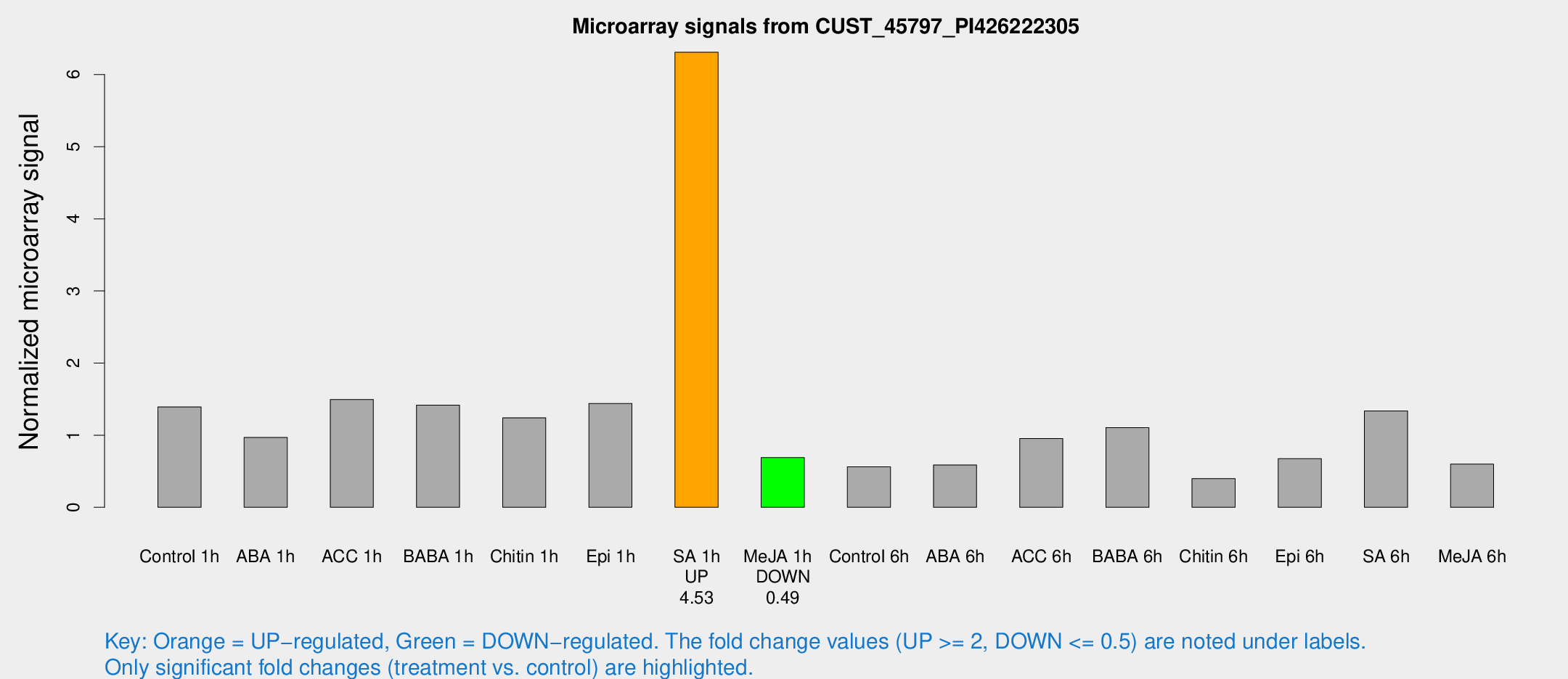
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1560.52 | 154.155 | 1.39172 | 0.0804031 |
| ABA 1h | 953.354 | 55.2412 | 0.967995 | 0.0559726 |
| ACC 1h | 1912.05 | 592.07 | 1.49479 | 0.451721 |
| BABA 1h | 1716.9 | 529.841 | 1.41552 | 0.409155 |
| Chitin 1h | 1248.52 | 101.809 | 1.24137 | 0.0717407 |
| Epi 1h | 1385.88 | 80.184 | 1.4394 | 0.0831698 |
| SA 1h | 7702.74 | 1823.2 | 6.30981 | 1.38003 |
| Me-JA 1h | 656.244 | 148.513 | 0.688887 | 0.0935133 |
| Control 6h | 651.083 | 122.865 | 0.560484 | 0.0667633 |
| ABA 6h | 792.115 | 258.882 | 0.587725 | 0.210373 |
| ACC 6h | 1218.77 | 70.6442 | 0.954133 | 0.179597 |
| BABA 6h | 1453.84 | 361.584 | 1.10463 | 0.23822 |
| Chitin 6h | 478.212 | 58.1445 | 0.398565 | 0.0567732 |
| Epi 6h | 881.827 | 188.402 | 0.674913 | 0.134361 |
| SA 6h | 1524.74 | 274.181 | 1.33549 | 0.11251 |
| Me-JA 6h | 684.082 | 124.626 | 0.5981 | 0.0595634 |
Source Transcript PGSC0003DMT400050270 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G14250.1 | +3 | 6e-64 | 210 | 98/268 (37%) | RING/U-box superfamily protein | chr3:4745963-4746958 REVERSE LENGTH=303 |
