Probe CUST_43518_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43518_PI426222305 | JHI_St_60k_v1 | DMT400065370 | TGCTGGGATTACAAAGTATATTGTACTTCCCCTCGCACATTTGAACGTTTATATATATGT |
All Microarray Probes Designed to Gene DMG400025409
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43518_PI426222305 | JHI_St_60k_v1 | DMT400065370 | TGCTGGGATTACAAAGTATATTGTACTTCCCCTCGCACATTTGAACGTTTATATATATGT |
| CUST_43522_PI426222305 | JHI_St_60k_v1 | DMT400065369 | ATCCCATGCCCATGGGCTACTTTACTGAAACAGTGATGGGAGCTTACAACTGCTTTGATT |
Microarray Signals from CUST_43518_PI426222305
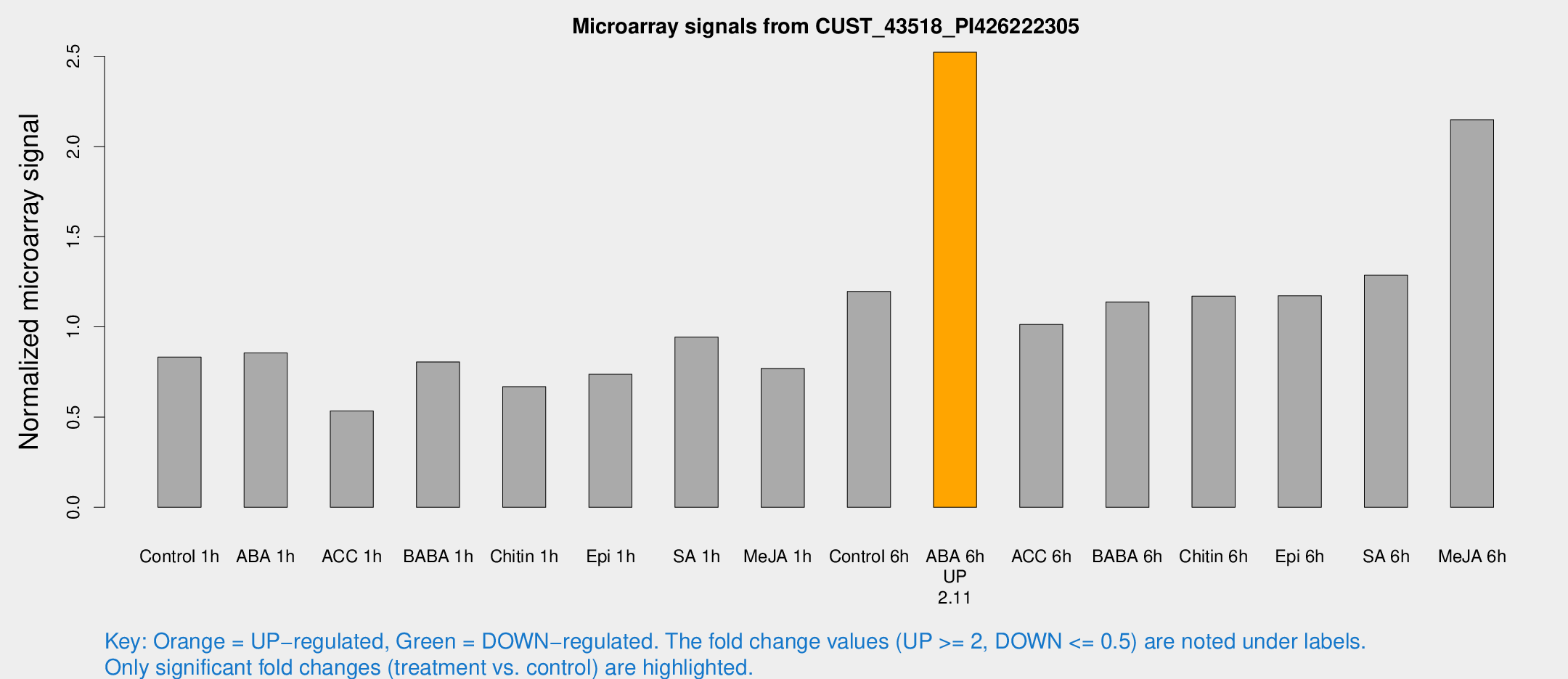
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1234.44 | 107.212 | 0.832264 | 0.0481076 |
| ABA 1h | 1152.77 | 196.795 | 0.855681 | 0.121032 |
| ACC 1h | 998.147 | 358.254 | 0.534545 | 0.270499 |
| BABA 1h | 1164.21 | 153.486 | 0.80553 | 0.0465824 |
| Chitin 1h | 915.079 | 162.62 | 0.668548 | 0.117233 |
| Epi 1h | 969.912 | 170.807 | 0.737518 | 0.133075 |
| SA 1h | 1431.49 | 91.8046 | 0.942823 | 0.0544812 |
| Me-JA 1h | 933.202 | 89.5993 | 0.769679 | 0.058953 |
| Control 6h | 1820.73 | 298.024 | 1.19675 | 0.110047 |
| ABA 6h | 3999.42 | 446.062 | 2.52233 | 0.165086 |
| ACC 6h | 1720.02 | 124.061 | 1.01339 | 0.0931542 |
| BABA 6h | 1906.76 | 232.62 | 1.13852 | 0.104477 |
| Chitin 6h | 1840.95 | 109.134 | 1.17072 | 0.0710138 |
| Epi 6h | 1961.03 | 168.602 | 1.17237 | 0.0677307 |
| SA 6h | 1910.15 | 250.679 | 1.28697 | 0.0743521 |
| Me-JA 6h | 3197.38 | 394.941 | 2.14818 | 0.124049 |
Source Transcript PGSC0003DMT400065370 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
